 A.
Overview
A.
Overview
As we've already mentioned, protein synthesis is fundamental to nearly everything a cell does. Protein channels are used to transport large molecules across the membrane. Almost all chemical reactions occuring in cells are catalyzed by protenaceous enzymes, including those involved in energy harvest, DNA replication, and cell division. Proteins perform important structural functions within cells and multicellular organisms, too; such as the histone proteins in chromosomes, the proteins in ribosomes, the collagen and elastin fibers that hold skin cells together, the collagen on which calcium and phosphate is deposited in bone, the protein myofibrils of actin and myosin in muscle cells, the neurotransmitters used for cell-cell communication between neurons, and the enzymes that digest food in the stomach and intestine of animals. So, proteins are fundamental to what cells and organisms ARE, structurally, and what they DO functionally. As you know, the genetic information determines the types of proteins a cell can make. The subset of proteins a cell actually DOES make, and the timing of WHEN they are made, is determined by what genes are "on" and what genes are "off" at a given time. This regulation of gene activity is ALSO co-ordinated by proteins - called transcription factors - that bind to DNA and promote or inhibit gene activity. So, proteins also regulate protein synthesis. Hopefully you see just how important proteins are to cells and organisms. So, the process of making these proteins is important, too.
 A.
Overview
A.
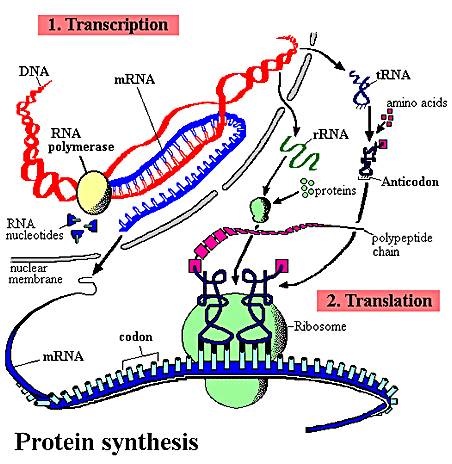
OverviewThe sequence of nitrogenous bases in a region of DNA is 'read' by a complex of enzymes that build a complementary strand of RNA. This process of reading DNA and making RNA is called 'transcription'. This is a great word for the process, as the message written in the language of nucleic acids is copied in essentially the same language - the language of nucleic acids. This RNA may be a recipe for a protein (m-RNA), or it may be an RNA that will act on its own as t-RNA, mi-RNA, si-RNA, or be complexed with proteins in the ribosome (r-RNA). Obviously, in "protein synthesis", only the m-RNA is read to make a protein. However, the other molecules all play a role. The sequence of nitrogenous bases in the m-RNA is then 'read' by a ribosome, which links a specific sequence of amino acids together into a protein based on that sequence of nitrogenous bases in the m-RNA. This process is called 'translation'. This is a great choice of a word, too. Here the sequence of information written in the language of nucleic acids is rewritten in a new language (hence, translation) - the language of amino acids.
Many of the initial RNA products have specific regions (introns) cut out of their sequence before they become functional. This step is known as "RNA processing" or "RNA splicing". Introns are present in nearly all eukaryotic RNA's, and are also in the DNA genes that encode them. Up until a few years ago, the only introns in prokaryotes had been found in t-RNA molecules of archaeans. More recently, however, introns have been found in m-RNA and r-RNA molecules of a few eubacteria and a few more archaeans. So, although they are rare in prokaryotes, we will describe a generic, simplified process of protein synthesis that includes introns and RNA processing.
In addition to splicing the RNA product of transcription, the initial protein product of translation may also be spliced and modified before it becomes functional. In eukaryotes, this protein processing often occurs in the Golgi apparatus.
The description presented here is a simple model of protein synthesis. You will learn more complex aspects of this process in Genetics.
1. Transcription:
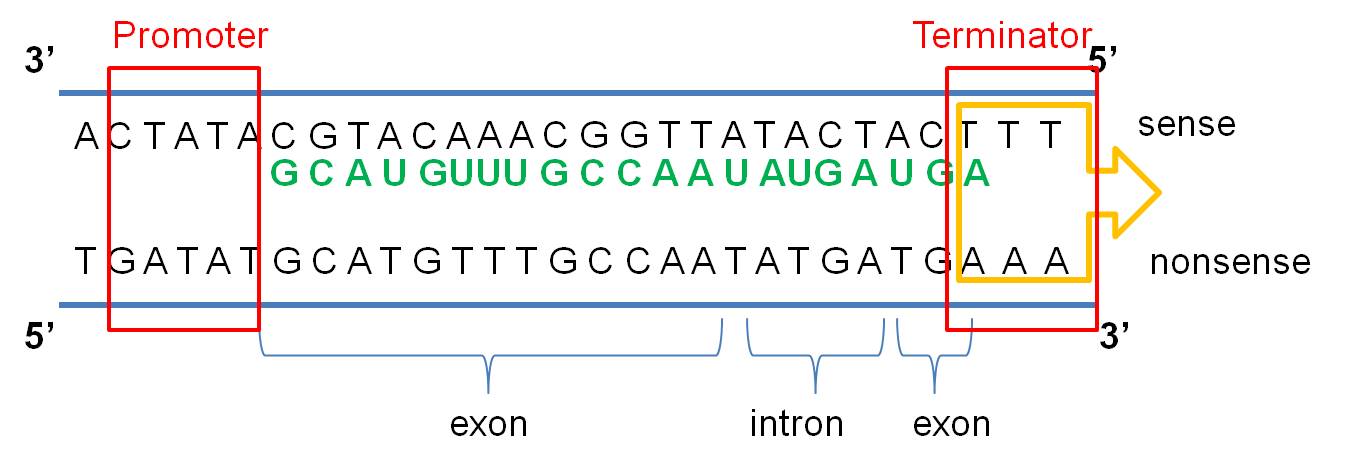 a. The message is on one strand of the
double helix - the sense strand: The DNA double helix is composed
of two anti-parallel complementary strands of DNA. Only one strand in a coding
region ("gene") is read; this strand carries a meaningful recipe that
"makes sense". This is called the "sense" strand. The other
strand, limited by complementarity, is not a meaningful message - it is the
"non-sense" (or "anti-sense") strand. Think about it this
way. Given a meaningful message of "C-A-T" (a small furry mammal),
the complementary strand is limited to the meaningless sequence of "G-T-A"
(????...). As the meaningful sequence gets longer, it is even LESS likely that,
just by chance, the complementary strand would be meaningful, too. Again, in
all eukaryotic genes and in some rare prokaryotic ones, there will be non-functional
"introns" interspersed throughout the meaningful message. The meaningful
parts are called "exons". The process of transcription is continuous,
so introns and exons get transcribed and these regions - if present in the DNA
- will also be present in the RNA product.
a. The message is on one strand of the
double helix - the sense strand: The DNA double helix is composed
of two anti-parallel complementary strands of DNA. Only one strand in a coding
region ("gene") is read; this strand carries a meaningful recipe that
"makes sense". This is called the "sense" strand. The other
strand, limited by complementarity, is not a meaningful message - it is the
"non-sense" (or "anti-sense") strand. Think about it this
way. Given a meaningful message of "C-A-T" (a small furry mammal),
the complementary strand is limited to the meaningless sequence of "G-T-A"
(????...). As the meaningful sequence gets longer, it is even LESS likely that,
just by chance, the complementary strand would be meaningful, too. Again, in
all eukaryotic genes and in some rare prokaryotic ones, there will be non-functional
"introns" interspersed throughout the meaningful message. The meaningful
parts are called "exons". The process of transcription is continuous,
so introns and exons get transcribed and these regions - if present in the DNA
- will also be present in the RNA product.
b. The cell 'reads' the correct strand based on the location of the promoter, the anti-parallel nature of the double helix, and the chemical limitations of the 'reading' enzyme, RNA Polymerase. RNA Polymerase binds to the DNA at a specific sequence next to the gene, called the 'promoter'. It binds in a specific way, so it is pointed towards the gene. RNA polymerase can only create a strand of RNA in the 5' to 3' direction, adding a new base to the free -OH group of the preceeding nucleotide on the chain. So, from its position at the promoter, looking down the two strands in the gene, the RNA Polymerase can only 'read' one strand - the DNA strand that is 3'-5'. It must create a strand that is anti-parallel to the DNA' template', and it can only bind nucleotides in 5'-3' direction. So, only the 3'-5' DNA strand is read in this region, and only one RNA strand, 5'-3' is made. It is important to appreciate that this relationship is 'local'. In another region of the DNA, the promoter may be on the other side of the gene, and the other strand may be read.
c.
Transcription ends at a sequence called the 'terminator'. These
regions have specific sequences that destabilize the attachment of the RNA Polymerase
to the DNA... it detaches and transcription stops. VIDEO
So, the process of transcription can be summarized like this: RNA Polymerase
binds at the promoter and reads the sense strand of DNA. The ploymerase links
together RNA nucleotides 5--> 3, in a sequence complementary to the DNA sense
strand. This process is continuous, so all DNA bases are 'read', including exon
and intron sequnces. This process continues until a terminator region is reached.
Reading this region destabilizes the RNA polymerase. It detaches from the DNA,
and transcription stops. All types of RNA (m-RNA, r-RNA, t-RNA) are made through
this process.
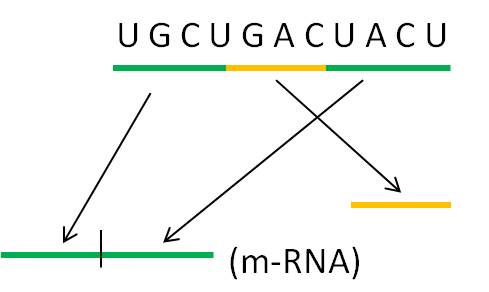 2.
Transcript Processing:
2.
Transcript Processing:
At this point in the process, the cell has read the gene and synthesized a complementary copy of strand of RNA. In all eukaryote sequences and many prokaryotic ones, this RNA molecule will have non-functional introns that need to be 'cut-out'. Enzymes cut the introns out and splice the ends together. In some cases, the introns catalyze their own excision - they are RNA molecules with enzymatic activity. These are one class of "ribozymes" - a very interesting class of molecules. There are other ribozymes that cleave other RNA molecules (not themselves) and others that catalyze other chemical reactions unrelated to RNA splicing.
In eukaryotes, the m-RNA, t-RNA and r-RNA is shunted through the nuclear membrane to the cytoplasm. In prokaryotes, there is no nucleus so the RNA is already in the cytoplasm. In all organisms, the r-RNA is complexed with proteins to form functional ribosomes. The t-RNA's bind specific amino acids.
3. Translation:
In this process, amino acids are linked together into a protein. The particular sequence of amino acids that are linked together is determined by the sequence of nitrogenous bases in m-RNA. This process occurs at the ribosome.
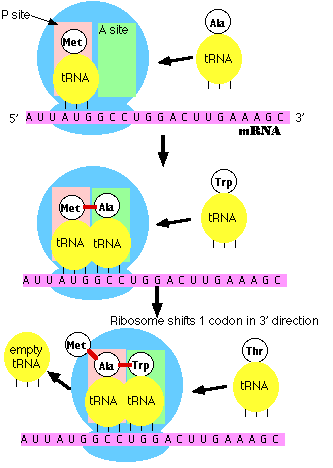 a.
m-RNA attaches to the ribosome at the 5' end.
The ribosome has two reactive sites. The RNA
moves through the ribosome until a specific sequence of nucleotides, AUG, is
positioned in the first site. Three base sequence in the m-RNA are called 'codons'.
This specific codon AUG, which starts the process of translation, is called
the 'start codon'. All proteins made by all life forms initially begin with
methionine, and use the codon AUG..
a.
m-RNA attaches to the ribosome at the 5' end.
The ribosome has two reactive sites. The RNA
moves through the ribosome until a specific sequence of nucleotides, AUG, is
positioned in the first site. Three base sequence in the m-RNA are called 'codons'.
This specific codon AUG, which starts the process of translation, is called
the 'start codon'. All proteins made by all life forms initially begin with
methionine, and use the codon AUG..
b. a specific t-RNA molecule, with a complementary UAC anti-codon sequence, binds to the m-RNA/ribosome complex. This t-RNA always carries the amino acid methionine. The genetic code describes the relationship between 3-base codons in m-RNA and the amino acids they code for.
c. Binding of the t-RNA to the first site opens a second site that reads the second 3-base codon (GCC in picture at right). Another t-RNA binds here - one with the specific anti-codon sequence (CGG). This t-RNA, with this anti-codon, always binds with the amino acid alanine.
d. Now a complex series of reactions occurs. Methionine is cleaved from its t-RNA and bound to alanine (this peptide bond between amino acids forms via dehydration synthesis). The t-RNA in position 1 vacates the site, and the t-RNA in site 2 moves to site 1. This is called a 'translocation reaction'. The next 3-base codon is positioned in the second site - ready to accept the next t-RNA/ammino acid complex (for tryptophan in the picture to the right).
e. Polymerization proceeds. This process continues down the m-RNA strand, reading the message one codon (3-base "word") at a time. For each codon, a specific amino acid is added to the chain. Thus, the nucleotide sequence in the m-RNA - copied from the nucleotide sequence in the DNA gene - determines the sequence of amino acids in the protein.
f. Termination. There are some codons that have no corresponding t-RNA molecule. When these codons enter the second site, no t-RNA/amino acid is added. When the ribosome translocates, no new amino acid is added and the chain is terminated. These particular codons that stop translation are called "stop codons".
4. Protein Processing:
The initial protein product usually needs to be modified to become functional. These modifications are termed "post-translational modifications". First of all, the methionine is usually cut off - this relieves an important constraint on the structure of functional proteins... functional proteins DON'T all start with methionine! Then, the protein may be spliced, or it may be bound with a sugar group (glycoprotein), lipid (lipoprotein), nucleic acid (nucleoprotein), or another protein (quaternary protein). In eukaryotes, much of this processing occurs in the Golgi apparatus.
Aside from somatic mutations, all the cells in a multicellular organism are genetically identical. So, the cells in your retina, bone, muscle, and stomach lining all contain the same genes. These cells perform different functions because they are reading different genes and making different proteins. Your muscle has the gene for rhodopsin (a photoreceptive pigment produced in the retina), but that gene is not transcribed in muscle cells. In contrast, retinal cells have the genes for the muscle proteins actin and myosin, but these genes are not transcribed. So, cell specialization and the developmental process by which cells specialize from the fertilized egg occurs by regulating this process of protein synthesis. Regulation can occur at each of the steps described above.
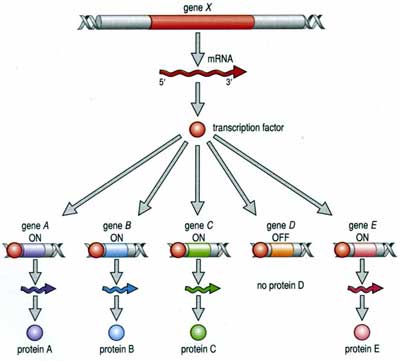 1.
Regulation of Transcription:
1.
Regulation of Transcription:
The process of transcription is regulated in several ways. First, the RNA polymerase can be blocked from the promoter. This can happen because the gene is bound to histones in a nucleosome, or is in a region of condensed 'heterochromatin', or because other proteins called 'transcription factors' have bound to the DNA - either at the promoter or between it and the gene, blocking the polymerase's route. However, the binding of other transcription factors can increase the affinity of the RNA polymerase for the promoter - increasing the probability of transcription. Again, these transcription factors are proteins encoded by other genes, and affected by other cellular processes. In this way, the action of a gene can be co-ordinated with the activity of other genes in a complex and interdependent manner. In addition, environmental cues from outside the cell can, through signal transduction, affect the activity of transcription factors and turn genes on or off. So, an organism can respond genetically to environmental cues.
2. Regulation of Transcript Processing:
The production of a protein can be affected at the processing stage. mi-RNA's and si-RNA's are small RNA molecules encoded by their own genes. These molecules can bind to m-RNA and effectively block correct splicing. This turns off production of the correct protein. In some cases, an initial m-RNA can be spliced two ways, creating two different functional products (and eventually two different proteins) depending on the pattern of cleavage. So, one gne may code for different proteins in different cells or tissue types.
3. Regulation of Translation:
One way that differential splicing can affect protein production is by changing the location of stop codons. For example, suppose a stop codon occurs at the beginning of an intron. Then, suppose that the intron is spliced incorrectly, after the location of this stop codon. Now, the resulting functional m-RNA has a stop codon where it didn't before; and translation will be terminated prematurely and no functional protein will be produced.
4. Regulation of Post-Translational Modification:
Initial protein products can be cleaved in different ways to produce different proteins, too.
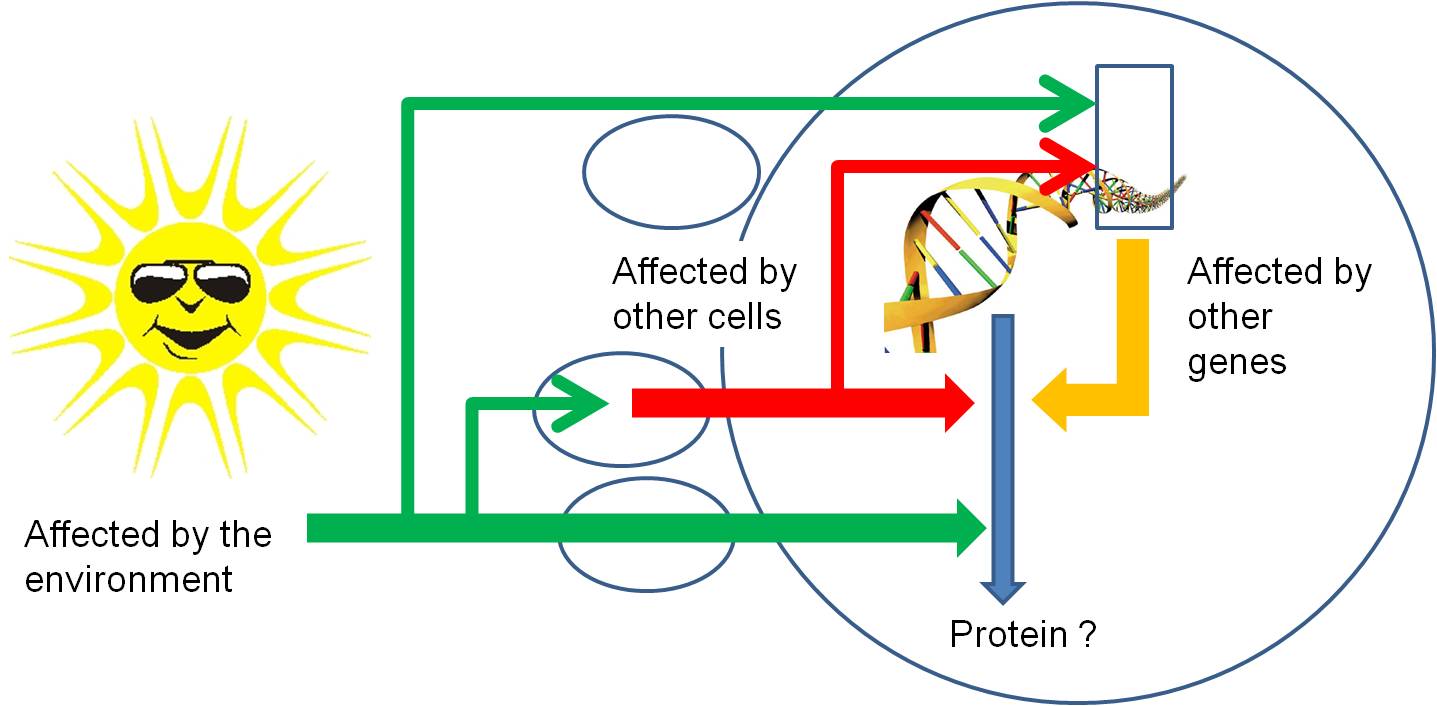
So, through all of these mechanisms, protein synthesis can be stopped or stimulated, and the product can be modified. Again, all of these regulatory pathways can be affected by environmental factors or the proteins or mi/si-RNA's produced by other genes. So, gene activity is affected by other things happening in the cell (turning other genes on and off) , in other cells of the organism (through the production of hormones that act as signal transducers), or environmental factors outside of the organism acting directly on this gene, on other genes in this cell, or on other cells..
STUDY QUESTIONS:
1. How does the polymerase "know" to read the sense strand? (several elements to this answer....)
2. What 'cues' determine where transcription will start and stop?
3. Consider the DNA double helix, below. Show what the RNA product will be after transcription. Remember - the RNA Polymerase lands on the promoter, and then reads one strand..... pick the correct one and transcribe it!
3'_________________________________5'.......
__________
| | | |
| | | |
| | | |
| |
|
|
A A T G C C C A T
T G G C A
|
|
PROMOTER
T T A C G G G T
A A C C G T
|
|
5'
| | | |
| | | |
| | | |
| | 3' ..... |
|
4. Describe the translocation reaction.
5. How is a polypeptide modified after translation to become a functional protein?
6. What are introns and exons, and how is protein synthesis modified to accomodate this structural change?
7. The process of protein synthesis and the universal genetic code provides one of the dramatic pieces of evidence of a common ancestry to all life. Explain why.