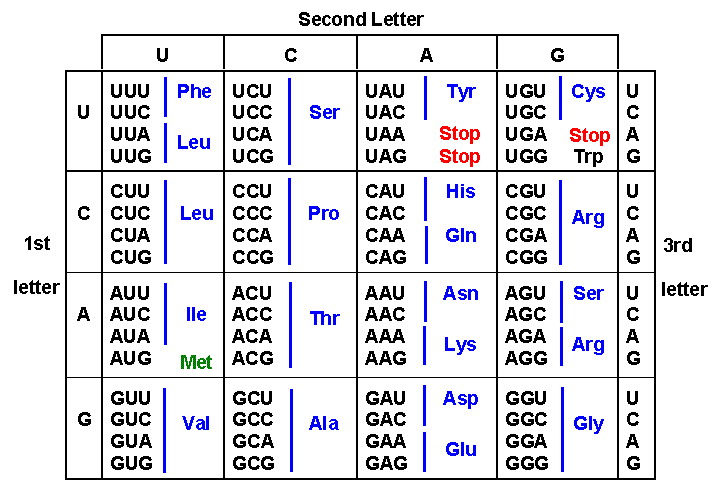
 The
genetic code describes the relationship between the three-base codon sequence
in an m-RNA molecule and the corresponding amino acid placed in a growing polypeptide
chain. The table is read in the following manner:
The
genetic code describes the relationship between the three-base codon sequence
in an m-RNA molecule and the corresponding amino acid placed in a growing polypeptide
chain. The table is read in the following manner:
 The
genetic code describes the relationship between the three-base codon sequence
in an m-RNA molecule and the corresponding amino acid placed in a growing polypeptide
chain. The table is read in the following manner:
The
genetic code describes the relationship between the three-base codon sequence
in an m-RNA molecule and the corresponding amino acid placed in a growing polypeptide
chain. The table is read in the following manner:
Suppose you wanted to find the amino acid coded for by the codon AAG. Begin on the left side of the chart, selecting the large row corresponding to the first letter - "A" (third major row). Now, move across that row to the cintersection with the third major row - corresponding the the second letter in the code ("A"). Four codons are presented in this box: AAU, AAC, AAA, and AAG. You can see that the AAG codon is associated with the amino acid 'Lysine'.
There are a few important characteristics of the code. First, there are "start" and "stop" codons. 'AUG' is the start codon, signified by the green coloring of the 'MET' (methionine) amino acid. The three stop codons are in red: UAA, UAG, and UGA.
Another important characteristic is that the code is 'unambiguous'. Each three base codon codes for only one amino acid. However, the code is also 'redundant', meaning that there are often more than one codon that codes (unambiguously) for a given amino acid. So, proline is encoded for by the codons CCU, CCC, CCA, and CCG. A pattern is fairly obvious - the third base in each codon is apparently less important in determining the amino acid than the first two. In fact, the binding between the third base in the anti-codon and the codon is 'sloppy', and mismatches here are tolerated. So, although the codon might be CCU, an incorrect t-RNA with the anti-codon GGG might bind (instread of the correct t-RNA with the anti-codon GGA). However, this incorrect t-RNA will still be carrying the correct amino acid proline. For some codon sets, the base at the third position is a little more important, but still not explicit. For example, tyrosine is specified by UAU and UAC. In these cases where only 2 (and not 4) codons specify one amino acid, the two codons that specifiy the same amino acid have either a purine or a pyrimide at the third position. In the previous example, tyrosine is specified by 'UA-pyrimidine'. So, it is obvious that the binding at the third position has rather variable chemcial specificity. In fact, only two amino acids are encoded by just one codon - methionine and tryptophan (trp). It is rathr easy to appreciate why the start codon would be unique.
| EXCEPTIONS TO THE UNIVERSAL GENETIC CODE | |||
| Organism | Normal codon | Usual meaning | New meaning |
| Mammalian mitochondria | AGA, AGG | Arginine | Stop codon |
| AUA | Isoleucine | Methionine | |
| UGA | Stop codon | Tryptophan | |
| Drosophila mitochondria | AGA, AGG | Arginine | Serine |
| AUA | Isoleucine | Methionine | |
| UGA | Stop codon | Tryptophan | |
| Yeast mitochondria | AUA | Isoleucine | Methionine |
| UGA | Stop codon | Tryptophan | |
| CUA, CUC, CUG, CUU | Leucine | Threonine | |
| Higher plant mitochondria | UGA | Stop codon | Tryptophan |
| CGG | Arginine | Tryptophan | |
| Protozoan nuclei | UAA, UAG | Stop codons | Glutamine |
| Mycoplasma capricolum bacteria | UGA | Stop codon | Tryptophan |
Table from: "Exceptions to the Genetic Code".
Even these exceptions 'prove the rule' of common descent, as major groups of organisms (mammals and plants) have the same exceptions across that group. In addition, all the changes (except the CU-block in yeast mitochondria) involve specificity in that labile third position. Finally it is interesting that the majority of these exceptions are in the mitochondrial genome of some eukaryotes. As endosymbiotic organelles, perhaps thay are not as reliant on their own protein synthesis as independent organisms and so have tolerated these mutations. The initial non-functional proteins might be compensated by proteins produced by the cell - allowing time for the new codon arrangement to evolve. So, although there are these few exceptions, the code is still considered 'universal'. An analogous evolutionary change would be the ñ in spanish or the way the "double LL" has come to represent the "Y" sound. These evolutionary novelties in that language do not falsify the broader, obvious relationships with french, nor their common ancestry from latin.