 |
 |
 |
 |
 |
Produced
by the Population Genetics and Evolution class, Furman University |
||||
 |
 |
 |
 |
 |
Produced
by the Population Genetics and Evolution class, Furman University |
||||
 |
The
Precambrian: The Last Universal Common Ancestor (LUCA) |
 |
It has been long supposed that if one were to trace down the total phylogenetic tree, eventually there would be a “root” organism: that is, the last universal common ancestor (LUCA) of all life. It is possible, though, that this was not a single organism, but rather a group of loosely associated, single cells with a very high rate of lateral gene transfer (Woese 1982). Perhaps Woese’s most potent argument against the idea of a single, organismic ancestor is this: for an organism to give rise to both Bacteria and Eukarya (the full sequence of an archaeal genome was not available at the time), it must at one time have been able to perform too enormous a variety of functions (those represented in those two domains) to have evolved in the proposed timeline of less than one billion years. Since Woese’s paper and the later sequencing of Methanococcus jannashii, it has been inferred through proteinomic analysis that the LUCA would have had to be even more complex than originally proposed, and that if it was a single organism, it was phylogenetically closest to Archaea and based upon analysis of archaeic, eukaryotic, and bacterial replication mechanisms, lacked its own replication mechanism (Kyrpides et al 1999). In a paper contradicting Kyrpides proposal of a lack of a replication mechanism, Ranea et al (2006) suggested that the LUCA (or comprising pool of cells) could, in fact, replicate itself. Further, analysis of protein domains shows that LUCA likely contained protein domains of almost all currently essential niches. Ranea et al make no comment on whether the LUCA was one or many organisms, but does provide the analogy of the LUCA to a human skeleton – having discovered the original “hard” and enduring characteristics merely serves to indicate how much “soft” and transient structure is absent from current knowledge. Page by Will Towler |
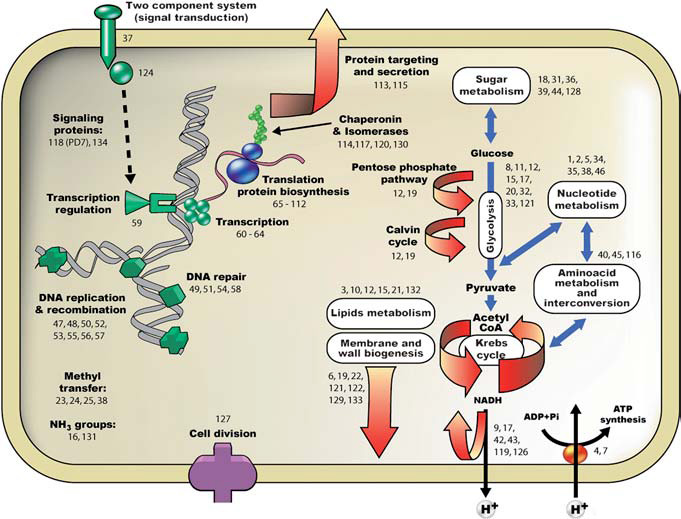 |
| “Schematic representation of cellular functions represented by the ancestral set of superfamilies The cellular and/or functional locations of the superfamilies’ domains are represented by numbers” (Renea et al 2006). | |
| Kyrpides
et al. 1999. “Universal Protein Families and the Functional
Content of the Last Universal Common Ancestor. “ . Accessed 18 Jan
2010.
Renea et al. 2010. “Protein Superfamily Evolution and the Last Universal Common Ancestor (LUCA). “ . Accessed 18 Jan 2010. Woese, C. 1982. “The universal ancestor. “ . 18 Jan 2010. |