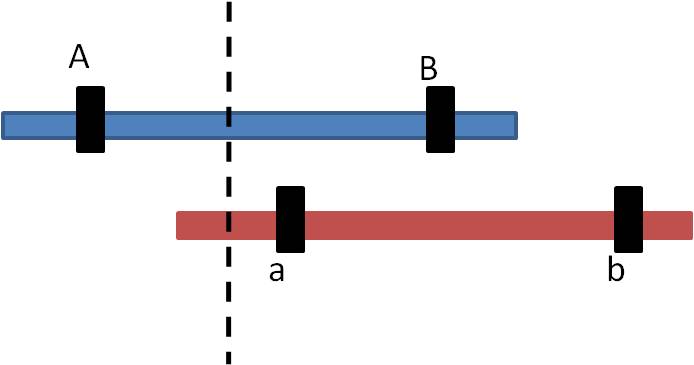
1. Mechanism #1: Unequal Crossing-Over:
a. process:
Sometimes during Prophase I of meiosis, the homologs do not line up evenly. If a cross-over event occurs as marked, then the pieces of DNA that are exchanged are not equivalent.

So, the products look like this:
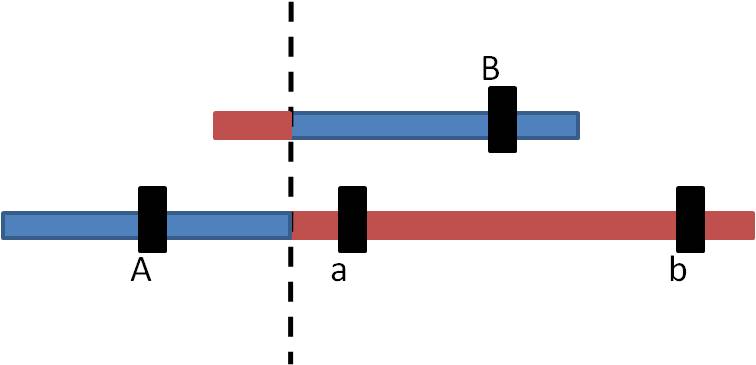
- The "A" locus has been DUPLICATED on the bottom chromosome, and DELETED from the top chromosome.
b. effects:
- Can be bad: DELETION of genes is usually bad... the gene probably did something useful, and now it is gone. At the least, organisms that inherit a deletion MUST express the gene at that lacus that they received from the other parent. So, deletions result in the increased frequency of expression of deleterious recessives inherited from the other parent. That's bad, too. (This is similar to why males expressed X-linked recessives more frequently than females - because they only get one X chromosome, so whatever is on it gets expressed. In that case, the pattern is true for ALL genes on the X chromosome. Here, with a deletion (that can occur on any chromosome), the effect is limited to the genes on the deletion region. But, whatever the organism inherits from the other parent must be expressed.)
DUPLICATION can also be bad. By having three active genes in an other wise diploid genome, the concentration of proteins produced by the duplicated gene may now be "out of whack" (a technical term) with the concentrations of other proteins. Essentially, it is similar to the effects of a trisomy, but at a smaller genetic scale.
- Can be good: Duplication of a gene can be good for two reasons. First, maybe more is better. As we mentioned before, there are many genes for r-RNA production, and many genes for melanin production. How did we get so many genes that are all the same, and code for the same thing? It would be VERY unlikely for random mutation, occurring in two different places in the genome, to create the same sequence of nitrogenous bases just by chance. So, how did it happen? Well, you just saw how - unequal crossing over. And if 'more is better', then organisms with more copies of a gene will be selected for, and these genomes will increase in frequency in the population.
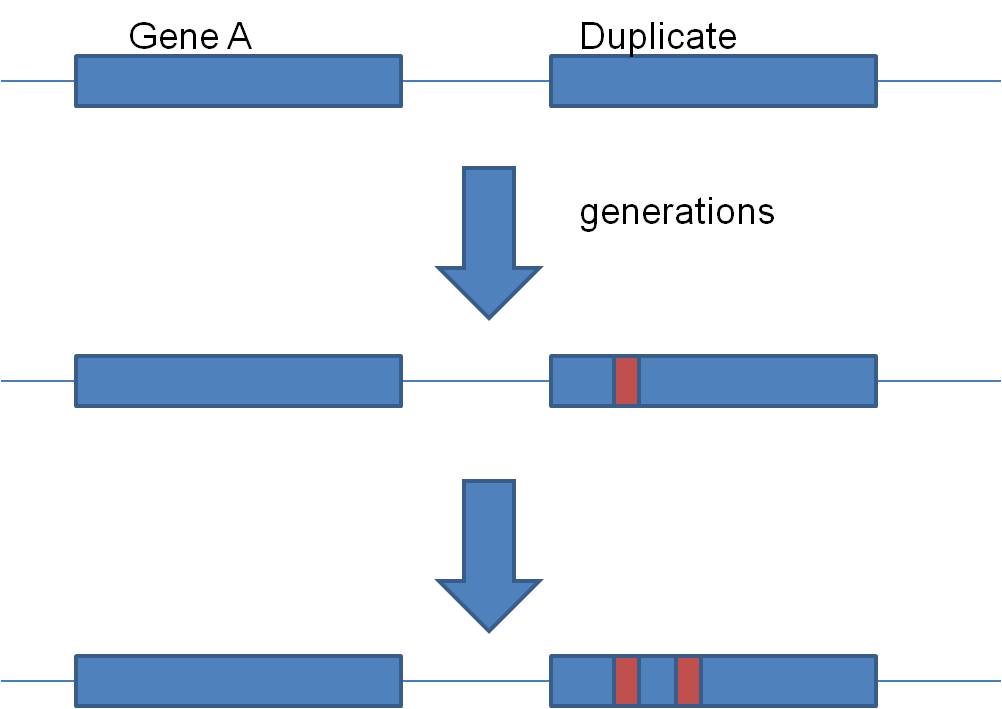 Second, gene duplication is an important source of EVOLUTIONARY NOVELTY - new,
useful variation. You see, it is easier to get something NEW and FUNCTIONAL
by modifying something that already works, rather than building it from scratch.
So, by duplicating a functional gene, evolution can now introduce mutations
into that "copy" and make something new. By tweaking that gene, we might
get something new that also works, but in a slightly different way. This would
produce a "family" of genes that all produce the same type of protein, but proteins
that work a bit differently. We call these families "gene families", and
they are quite common. A classic example is the "globin family" of genes.
This includes the alpha, beta, gamma, and myo- globin genes. They are
all similar enough to suggest common ancestry from one ancestral gene, yet different
enough to do slightly different things. And they are all similar enough
in nucleotide sequence that their origin by duplication and subsequent slight
modification is strongly indicated.
Second, gene duplication is an important source of EVOLUTIONARY NOVELTY - new,
useful variation. You see, it is easier to get something NEW and FUNCTIONAL
by modifying something that already works, rather than building it from scratch.
So, by duplicating a functional gene, evolution can now introduce mutations
into that "copy" and make something new. By tweaking that gene, we might
get something new that also works, but in a slightly different way. This would
produce a "family" of genes that all produce the same type of protein, but proteins
that work a bit differently. We call these families "gene families", and
they are quite common. A classic example is the "globin family" of genes.
This includes the alpha, beta, gamma, and myo- globin genes. They are
all similar enough to suggest common ancestry from one ancestral gene, yet different
enough to do slightly different things. And they are all similar enough
in nucleotide sequence that their origin by duplication and subsequent slight
modification is strongly indicated.
There is ANOTHER reason why duplication is an important source of evolutionary novelty, and it relates to an "intelligent design" criticism of evolution. Proponents of intelligent design and creationism suggest that evolution by gradual change is impossible if the intermediate steps are non-functional. For instance, how could the beta-globin gene "evolve from" an alpha globin gene by a gradual process if the intermediate steps are non functional? You see, here's the problem:
 Suppose alpha
globin differs from beta globin by 5 amino acids (I have no idea how many -
this is just an example). In order for the beta globin gene to "evolve",
5 different mutations in the DNA would have to occur, in order to code for five
new amino acids. Now, it is VERY unlikely that all five of those particular
mutations would occur at the same time - mutations are very rare. BUT
(argue the creationists), that's exactly what would have to happen for beta
globin to evolve from alpha globin. Otherwise, they argue, ff we have
one mutation at a time, the alpha globin is turned off and we have NO alpha
globin and NO beta globin - a non-functional intermediate that should be selected
against.
Suppose alpha
globin differs from beta globin by 5 amino acids (I have no idea how many -
this is just an example). In order for the beta globin gene to "evolve",
5 different mutations in the DNA would have to occur, in order to code for five
new amino acids. Now, it is VERY unlikely that all five of those particular
mutations would occur at the same time - mutations are very rare. BUT
(argue the creationists), that's exactly what would have to happen for beta
globin to evolve from alpha globin. Otherwise, they argue, ff we have
one mutation at a time, the alpha globin is turned off and we have NO alpha
globin and NO beta globin - a non-functional intermediate that should be selected
against.
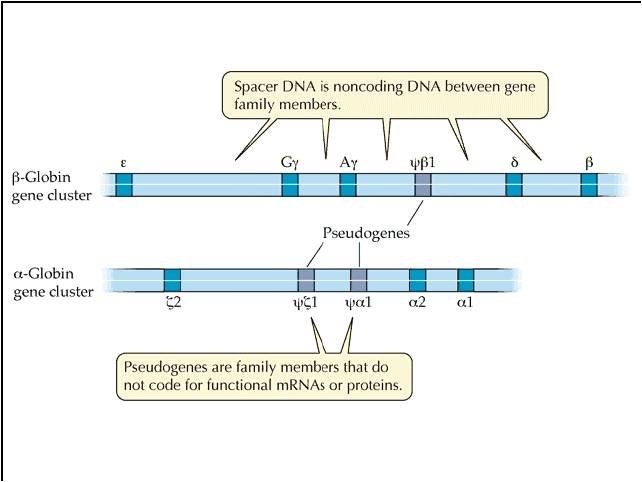
BUT, duplication solves this apparent dilemma. If the alpha gene duplicates, there are now TWO alpha globin genes. Suppose the first mutation occurs in one, and shuts that gene off. So what? The other gene is still right there, cranking out the functinal alpha globin protein. So, the organism with the non-functional duplication is NOT selected against. Rather, they pass their chromosome on to offspring, with the functional and non-functional genes. Generations later, the 2nd, 3rd, 4th, and 5th mutations can accumulate. When the 5th happens, the product is functional beta globin.... and now the organisms that have inherited this mutation have both functional alpha and functional beta globin. Gene duplication is a VERY important mechanism of introducing evolutionarily SIGNIFICANT variation, because it maintains the original gene and its function, while allowing mutations to "play" with the other gene. Mutations that turn that genes off are not selected against, but mutations that improve the gene/protein can still have a positive effect and be selected for. In fact, many regions of non-coding DNA in our genome are "pseudogenes".... regions of non-functional DNA that bear a remarkable similarity in nucleotide sequence to a functional gene. The hypothesis is that they represent a duplicated gene that has since been turned off by mutation.
There is also another bit of evidence here. We often see members of gene families
in the same region of a chromosome - just as we would expect from unequal crossing-over
that places a duplicate next to its homologous gene. So, both the sequence similiarity
and the spatial proximity that we often see in gene families is strong evidence
that unequal cross-over and gene duplication have been important sources of
new useful genes.
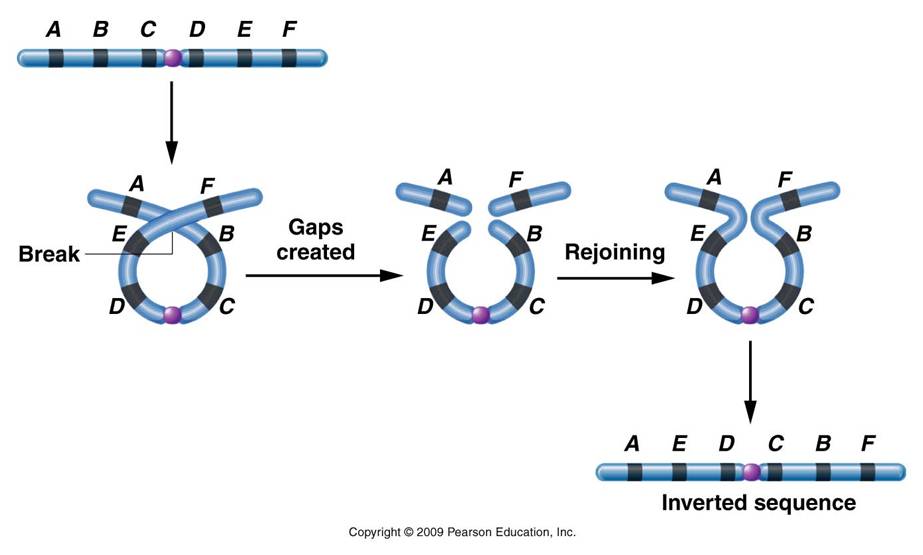 2.
Mechanism #2: Inversions - changing gene order
2.
Mechanism #2: Inversions - changing gene orderChromosomes can coil up and cross-over with themselves; inverting a region of genes. The most significant effect that this has on heredity is that subsequent crossing over events between this inversion chromosome and its correctly-ordered homolog now result in non-functional products. The chromosomes that result from crossing over have deletions and duplications. So, the only functional gametes that this organism makes are the parental types - those preserving the arrangement of alleles on each chromosome - including the new arrangment in the inversion region. So, inversions 'stabilize' a set of alleles, preventing their recombination through crossing over. As such, groups of alleles are inherited as a unit, and can be selected for or against as a group. This process can stabilize beneficial combinations of alleles.
 3.
Mechanism #3 - Translocation: moving genes to different chromosomes
3.
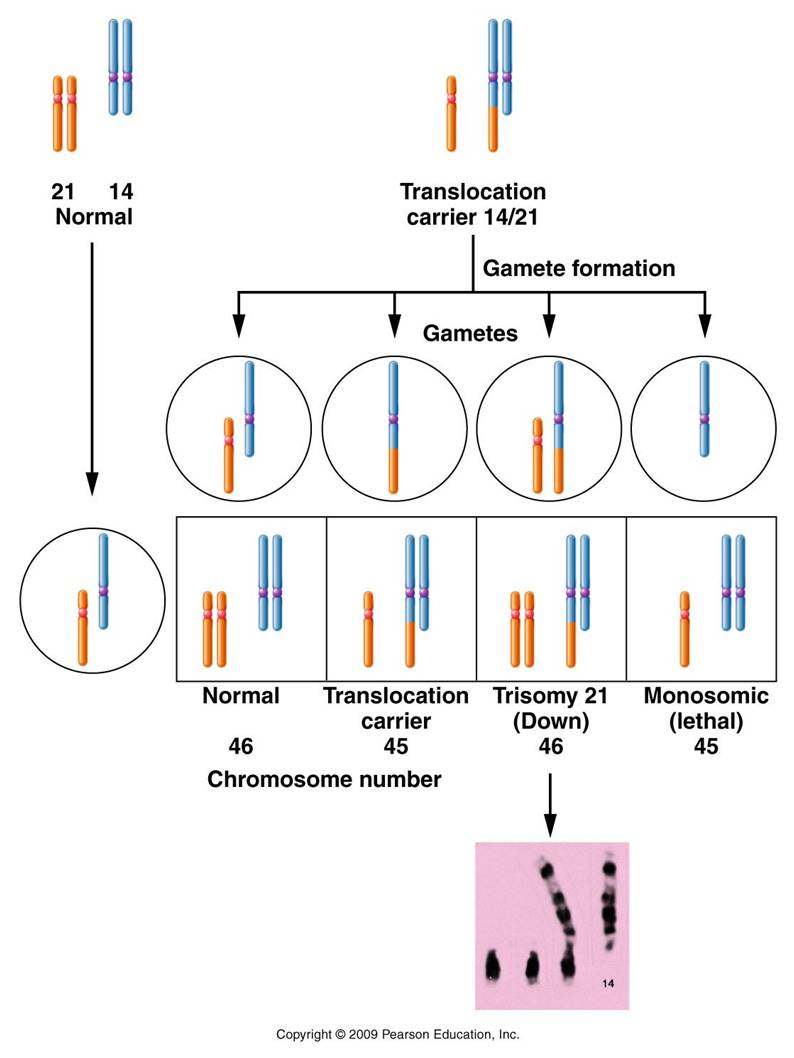
Mechanism #3 - Translocation: moving genes to different chromosomes
this is an exchange (or unidirectional transfer) between NON-HOMOLOGOUS chromosomes.
An example is Translocation Down's syndrome. This is where one of the #21 chromosomes gets 'stuck' (translocates) to a #14 chromosome. The individual with this genotype is unaffected - they have the correct DNA content, it's just in a new spatial configuration. But when this individual produces gametes, problems can occur. So... the person has a normal 14, a normal 21, and the translocation chromosome (14+21). The 14's will pair and separate during Meiosis I. If the free 21 goes with the normal 14, you get a normal gamete. But the free 21 could also go with the 14+21 chromosome - resulting in a gamete with two 21's, and another cell with only a 14 (zygote aborted). Fertilization with a normal gamete will produce a zygote with 2 #21's, a free 14, and the 14+21. Two doses of 14 (correct), but 3 doses of 21 (causing Down's syndrome). Trisomy 21 is not heritable. This means that trisomy 21 does not run in families; it is caused by a random non-disjunction error. Translocation Down's, however, IS heritable. So, think about it this way. Suppose you have a sibling with Down's syndrome. If their Down's is caused by trisomy 21, then you would have no greater chance than anyone else in the population of having a child with Down's syndrome. However, if your sibling has translocation Down's, then there is a 50% chance that you - even though you are normal phenotypically - are a carrier of the translocation chromosome. As such, there is a 1/3 chance that you could have a child with translocation Down's.
1. Mechanism #1: Crossing Over within eukaryotic genes can produce new combinations of exons, and new alleles:
- In eukaryotes (and some prokaryotes) with the exon-intron structure of their genes, crossing over WITHIN A GENE, in non-coding intron regions, can create new combinations of exons. This may be a particularly productive way of creating new functional alleles. Why? Well, consider two alleles, A and a, that both function. For simplicity, suppose they are composed of 3 exons each:
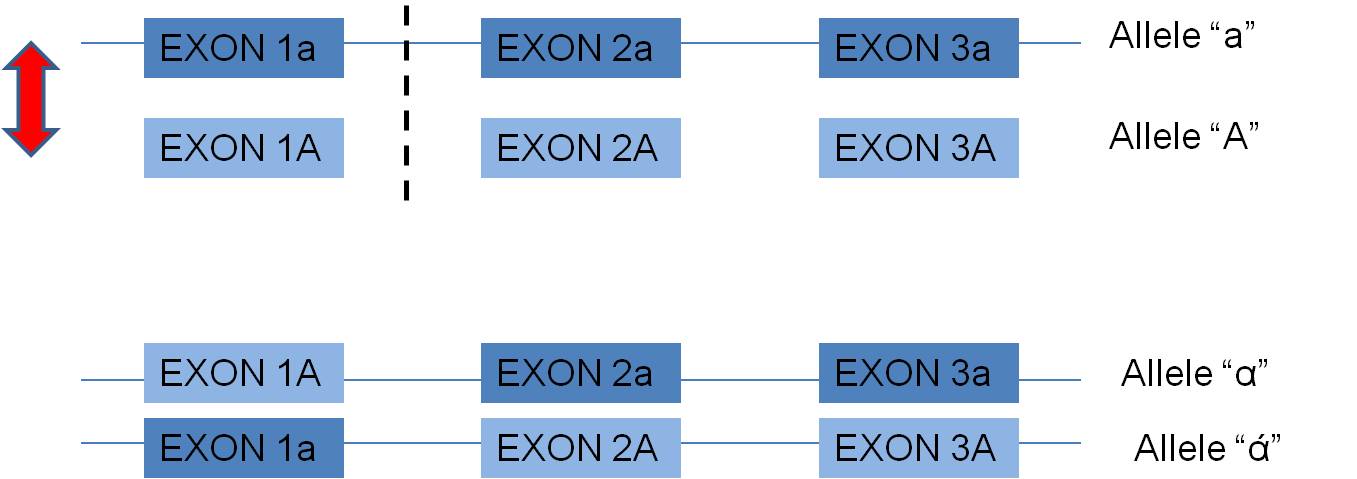
Now, the exons already function with one another. A cross-over between the first and second exon may produce new combinations of exons that also have a high probability of functioning, but maybe in a slightly new way. In fact, many genes across the genome seem to share particular exons that encode for a particular functional domain of proteins. It is far more likely that these are homologous similiaries arising from "exon shuffling" than the independent but parallel creating of these identical sequences by mutation.
2. Mechanism #2: Point Mutations - changes in nucleotide sequence
a. Addition/deletion:
When DNA
is being replicated, the DNA polymerase may erroneously "skip" a base, or erroneously
add an extra base into the newly forming strand. If this is the sense
strand that will later be transcribed, this will cause the addition or deletion
of a base in the m-RNA. This change will "shift" the 3-base reading frame
during translation, changing every codon downstream: 
So, from the point of the addition or deletion, every amino acid that follow will change. So, these "frameshift" mutations usually have a deleterious effect because they change a whole bunch of amino acids.
b. Substitutions:
- during DNA replication, an improper base can be substituted while the new strand is being synthesized. Many of these errors are corrected during 'DNA proof-reading' and repair that occurs in G1 and G2, but some (1 in a million) slip by. This change in the DNA (if in an exon) would change one base in the m-RNA, and change one m-RNA codon. This could change an amino acid - but if you look at the genetic code you will see that most AA's are encoded by more than one three base codon. So, for instance, CUU, CUA, CUG, and CUC all code for the amino acid "leucine". So, a change in the third position of the DNA sequence encoding these codons would have NO EFFECT on the amino acid:
Original DNA = GAT ....... original RNA = CUA ....... amino acid = leucine
mutant DNA = GAA ........ mutant RNA = CUU ...... amino
acid is STILL LEUCINE
So, some mutations are "silent", even if they occur within an exon of a gene.
However, other mutations can change the amino acid:
Original DNA = GAT ....... original RNA = CUA ....... amino acid = leucine
mutant DNA = GTT ........ mutant RNA = CAA ...... amino
acid is GLYCINE

The smallest change that is possible in DNA is a single nucleotide, and this may cause the smallest change possible in a protein - a change in a single amino acid. These small changes may be deleterious, neutral, or beneficial. And, AS YOU KNOW, the value of a gene may depend on the environment.
Consider the sickle cell anemia example that we discussed before. Hemoglobin consists of two alpha globin molecules and two beta globin molecules. The allele for the normal beta globin chain contains 146 amino acids, with glutamine as the 6th amino acid. The sickle cell hemoglobin contains the SAME alpha chains as normal hemoglobin, but a mutant form of the beta chain. In the mutant form, the 6th amino acid is valine. This difference in one amino acid is caused by a change in one DNA nucleotide. As we have discussed, it is beneficial in the tropics but deleterious in the temperate zone.
We began the unit with Darwin's model of evolution:
Sources of variation
Agents of change
unknown
Natural selection.
As a consequence of our modern understanding of heredity and genetics, we have learned quite a bit about variation AND evolution. Our model, a this point in the class is:
Sources of variation
Agents of Change
MUTATION:
-New Genes:
Natural Selection
point mutation
Mutation (polyploidy can make new species)
exon shuffling
RECOMBINATION:
- New Genes:
crossing over
-New Genotypes:
-crossing over
- independent assortment
Study questions:
- Alpha and beta globin molecules are VERY similar to one another in nitrogenous base sequence. What does this suggest about where they came from? How might this have happened?
- Describe two reasons why duplications may be advantageous.
- How are new genes produced? List two ways (in eukaryotes).
- How is translocation Down's caused?
- Why are substitution mutations likely to have a less pronounced effect tha addition or deletion mutations?
- Outline the causes of genetic variation.