 A.
DNA and RNA Structure
A.
DNA and RNA Structure
 A.
DNA and RNA Structure
A.
DNA and RNA Structure
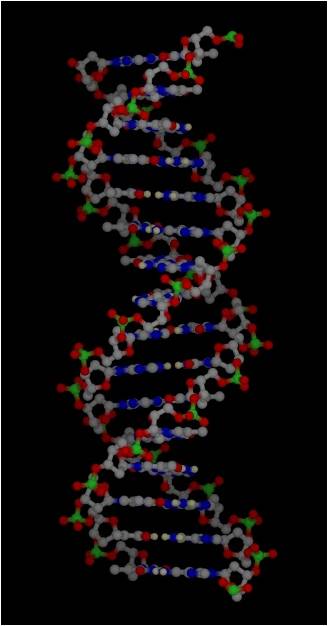
DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are nucleic acids - polymers consisting of a linear sequence of linked nucleotide monomers. We will describe the structure of the monomers first, and then describe how they are linked into linear polymers. Finally, we will describe the double-stranded structure of ds-DNA.
1. The monomers are "nucleotides"
three components:
- Pentose (5 carbon) sugar: either ribose (RNA) or deoxyribose (DNA). The carbons are numbered clockwise. The difference between the sugars is that ribose has an -OH group on the 2' carbon, whereas deoxyriboes has only 2 H groups and thus is "deoxygenated" relative to ribose. BOTH sugars have an -OH group on the 3' carbon, which will be involved in binding. The 5' carbon is a sidegroup off the ring.
- Nitrogenous Base: each nucleotide has a single nitrogenous base attached to the 1' carbon of the sugar. This nitrogenous base may be a double-ringed structure (purine) or a single ringed (pyrimidine) structure. The purines are adenine (A) and guanine (G). The pyrimidines are thymine (T), cytosine (C), and uracil (U). DNA nucleotides may carry A, G, C, or T. RNA nucleotides carry either A, G, C, or U.
- The third component of a nucleotide is a phosphate group, which is attached to the 5' carbon of the sugar. When a nucleotide is incorporated into a chain, it has a single phosphate group. However, nucleotides can occur that have two or three phosphate groups (dinucleotides and trinucleotides). ADP and ATP are important examples of these types of molecules. In fact, the precursors of incorporated nucleotides are trinucleotides. When two phosphates are cleaved, energy is released that can be used to add the remaining monophosphate nucleotide to the nucleic acid chain.
2. Polymerization is by 'dehydration synthesis'
As with all other classes of biologically important polymers, monomers are linked into polymers by dehydration synthesis. In nucleic acid formation, this involves binding the phosphate group of one nucleotide to the -OH group on the 3' carbon of the existing chain. For the purposes of seeing how this reaction works, we can envision an H+ on one of the negatively charged oxygens of the phosphate group. Then, a molceule of water can be removed from these two -OH groups, leaving an oxygen binding the sugar of one nucleotide to the phosphate of the next.
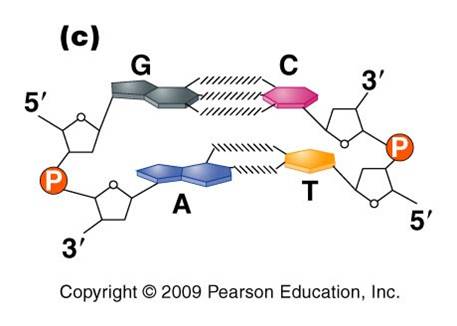
This creates a 'dinucleotide'. It has a polarity/directionality; it is different at its ends. At one end, the reactive group is the phosophate on the 5' carbon. This is called the 5' end of the chain. At the other end, the reactive group is the free -OH on the 3' carbon; this is the 3' end of the chain. So, a nucleic acid strand has a 5' - 3' polarity.
 3.
Most DNA exists as a 'double helix' (ds-DNA) containing two linear nucleic acid
chains.
3.
Most DNA exists as a 'double helix' (ds-DNA) containing two linear nucleic acid
chains.
a. the nitrogenous bases on the two strands are 'complementary' to each other, and form weak hydrogen bonds between them. A always pairs with T, and C always pairs with G. As such, there is always a double-ringed purine pairing with a single-ringed pyrimidine, and the width of the double-helix is constant over its entire length.
b. the two strands (helices) are anti-parallel: they are arranged with opposite polarity. One strands points 5' - 3', while the other points 3' - 5'. The direction of the pentose sugars and the type of reactive group at the ends of the chains show this relationship.
4. RNA performs a wide variety of functions in living cells:
a. m-RNA (for "messenger") is the copy of a gene. It is the sequence of nitrogenous bases in m-RNA that is actually read by the ribosome to determine the structure of a protein.
b. r-RNA (for "ribosomal") is made the same way, as a copy of DNA. However, it is not carrying the recipe for a protein; rather, it is functional as RNA. It is placed IN the Ribosome, and it helps to ‘read’ the m-RNA.
c. t-RNA (for "transfer") is also made as a copy of DNA, but it is also functional as an RNA molecule. Its function is to bind to a specific amino acid and incorporate it into the amino acid sequence as instructed by the m-RNA and ribosome.
d. mi-RNA (micro-RNA) and si-RNA (small interfering RNA) bind to m-RNA and splice it; inhibiting the synthesis of its protein. This is a regulatory function.
e. sn-RNA (small nuclear RNA) are short sequences that process initial m-RNA products, and also regulate the production of r-RNA, maintain telomeres, and regulate the action of transcription factors. Regulatory functions.
 B.
Chromosome Structure
B.
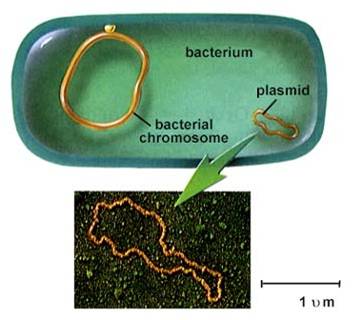
Chromosome Structure1. Prokaryotes
- usually one circular chromosome, tethered to the membrane, with some associated,
non-histone proteins.
2. Eukaryotes
– usually many linear chromosomes, highly condensed with histone proteins into several levels of structure.
Level 1: ds-DNA is wrapped around histone proteins, creating the “beads on a string’ level of organization.
Level 2: string is coiled, 6 nucleosomes/turn (solenoid)
Level 3: the coil is ‘supercoiled’
Level 4: the supercoil is folded into a fully condensed metaphase chromosome
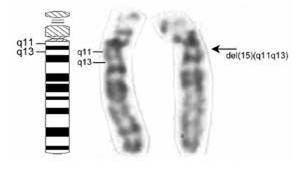 To
read a gene, the chromosome must be diffuse (uncondensed) in that region. Even
when condensed, these ‘euchromatic’ coding regions are less condensed
and more lightly staining than non-coding regions.
To
read a gene, the chromosome must be diffuse (uncondensed) in that region. Even
when condensed, these ‘euchromatic’ coding regions are less condensed
and more lightly staining than non-coding regions.
DNA that has few genes can remain
condensed and closed (heterochromatic), and appears as dark bands on condensed
chromosomes.
Study Questions:
1) Diagram the parts of an RNA nucleotide.
2) Show how two nucleotides are linked together by dehydration synthesis reactions.
3) Why does the purine - pyrimidine structure relate to the complementary nature of double-stranded DNA?
4) Draw a DNA double helix, showing three base pairs and the antiparallel nature of the helices.
5) Describe the higher levels of eukaryotic chromosome structure, including the terms nucleosome and solenoid.
6) What are two differences between euchromatin and hetochromatin?