Heredity, Gene Regulation, and Development
I. Mendel's Contributions
II. Meiosis and
the Chromosomal Theory
D. The Process of Meiosis
1. Overview
of the Process:
Meiosis has two cycles of division, preceeded by an interphase containing a
G1, S, and G2 phase. So, the cell entering meiosis has replicated chromosomes.
The first cycle is called Meiosis I, or the reduction cycle. A diploid
cell containing two sets of replicated chromosomes divides and produces cells
that are haploid, containing one set of replicated chromosomes. The second cycle
is Meiosis II, also called the division cycle. In this cycle, the haploid
cells divide in a manner much like mitosis. In other words, the sister chromatids
of each replicated chromosome are separated, but there is no change in ploidy.
The haploid cell with replicated chromosomes divides into haploid cells with
unreplicated chromosomes .
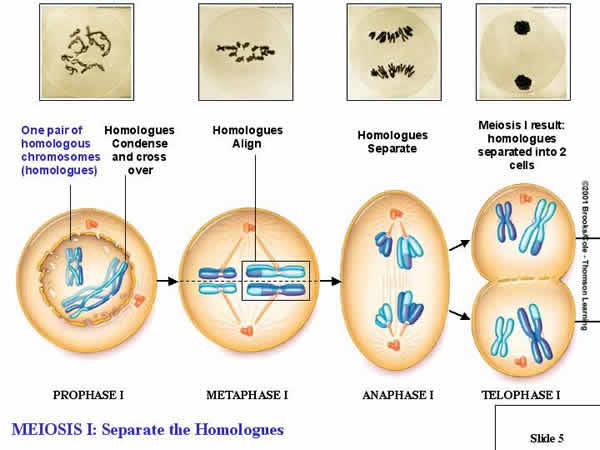 2.
Meiosis I (Reduction)
2.
Meiosis I (Reduction)
a.
Prophase I:
- The chromosomes condense as
paired homologs.
DEFINITION: Homologous
chromosomes govern the same suite of traits, but may affect those traits in
different ways. So, if there is a chromosome with a gene for purple flower color
at a particular spot (locus), then it's HOMOLOG will also have a gene for flower
color at that same spot... but it might be a different ALLELE for flower color
(white).
- As condensation occurs, replicated,
homologous chromosomes pair up. Each replicated chromosome has two chromatids,
so the pair of homolos for a structure with four chromatics - a 'tetrad'. Piece
of chromosome can be exchanged between homologs. This is called 'crossing over'
. This creates new combinations of genes on chromosomes. we will consider this
in more detail in a couple days.
- Asd the spindles join to paired
homologs, the spindle fibers from each pole can only attach to ONE homolog or
the other.
b.
Metaphase I:
- THIS IS THE MOST IMPORTANT
STEP, AND THE THING THAT DISTINGUISHES MEIOSIS FROM MITOSIS... Homologous
pairs line up on the metaphase plate (NOT in single file as in mitosis, but
as PAIRS).
c.
Anaphase I:
- Whole, replicated chromosomes
are drawn to each pole by the spindle fibers. So, if the 'A' chromosome goes
to one pole, the 'a' chromosome goes to the other.
d.
Telophase I:
- The cytoplasm divides; each new
cell has only ONE chromosome from each homologous pair. If the cell started
with 4 chromosomes (2n = 4), NOW each daughter cell has only 2 chromosomes
(1n = 2). This is REDUCTION, and it is the most important part of Meiosis.
It occurs because homologs pair up in metaphase I, rather than lining up in
single file as in mitosis.
3. An optional interphase:
The transition from meiosis I to meiosis II cay vary between species. In some
species, the nuclear envelope reforms around the chromosomes in each nucleus.
There may even be an interphase in which the chromosomes decondense, protein
synthesis occurs, and the cells grow. However, no S phase occurs in this interphase
- the chromosomes are already in their replicated state. If this interphase
does occur, then Prophase II begins with the condensation of the chromosomes
and the breakdown of the nuclear membrane.
In other species, the cells proceed directly from Telophase I to Prophase II;
the chromosomes are already condensed and the nuclear membrane is already broken
down. In this case, the only thing that happens is that a new spindle forms,
attaching each chromosome to both poles of the cell.
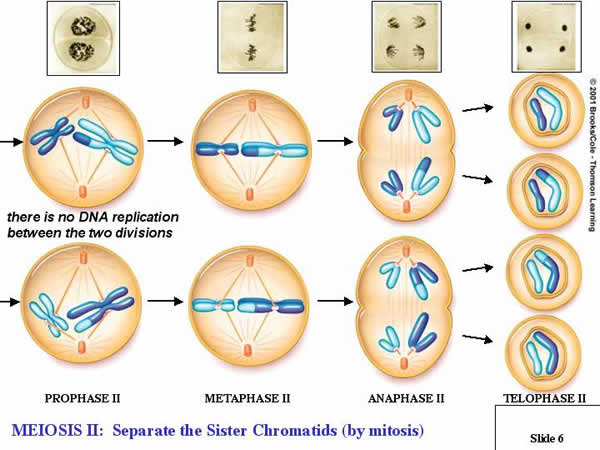 4.
Meiosis II: (Division):
4.
Meiosis II: (Division):
a.
Prophase II:
Depending on the transition, events
proceed such that the new spindle apparatus attaches condensed chromosomes to
both poles of each cell produced in the reduction cycle (Meiosis I).
b.
Metaphase II:
Replicated chromosomes line up in
single file on the metaphase plate, in the middle of each cell.
c.
Anaphase II:
The chromatids in each replicated
chromosome are drawn to opposite poles in each cell.
d.
Telophase II:
The nuclear membranes reform while
cytokinesis divides the cells.
video
5. Modifications in Anisogamous Species:
In isogamous species and in homosporous plants (that produce one type of spore
by meiosis), the reproductive cells produced by meiosis are all the same size.
In this case, the process works as described above, and one parental cell produces
four equal-sized daughter cells.
In anisogamous species (and in heterosporous plants that produce different
sized spores by meiosis), the reproductive cells produced by male and female
organisms vary in size. Although the division of the genetic material occurs
in the same way (creating daughter cells that a have exactly half of the genetic
information of the parent cell), the division of the cytoplasm differs in these
processes.
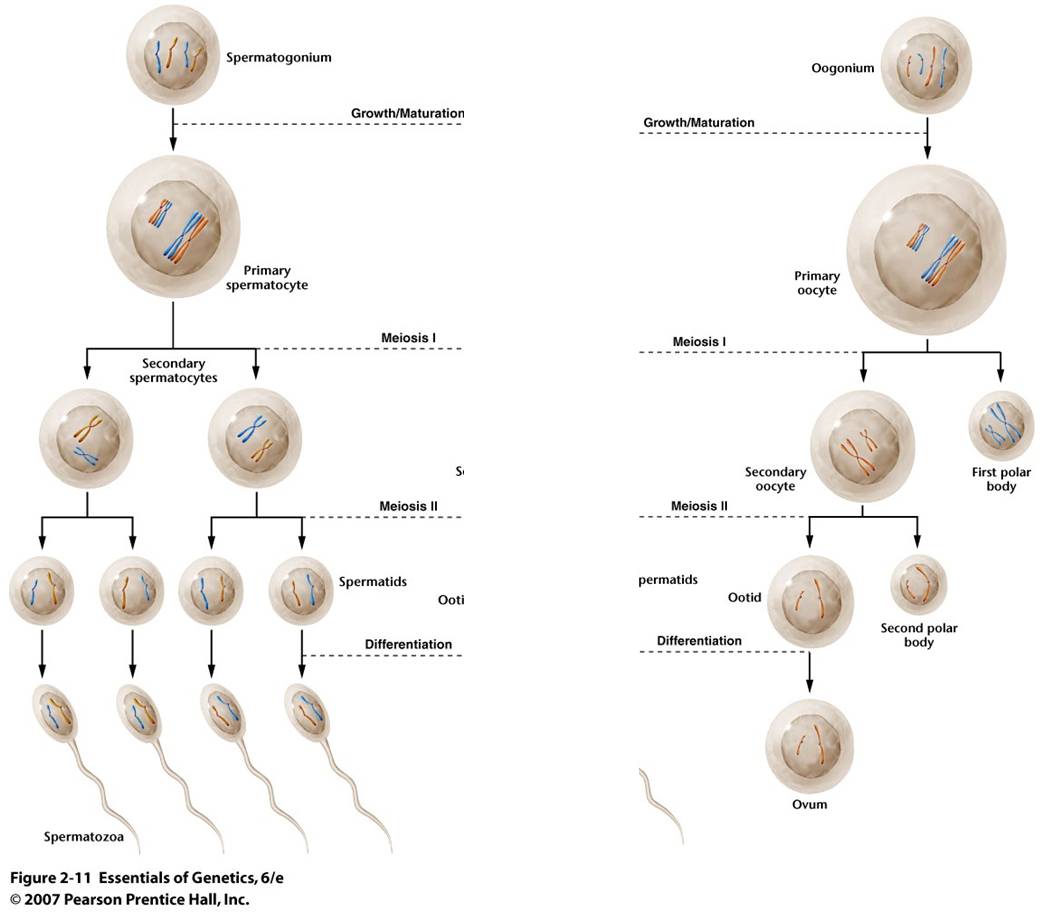 a.
Spermatogenesis:
a.
Spermatogenesis:
TIn sperm
production, the division of the genetic information is EVEN, and the division
of the cytoplasmic material is EVEN. So, Spermatogenesis produces four small
functional sperm cells from one initial diploid cell.
b. Oogenesis:
Egg production
is a little different. The division of the genetic information is EVEN, just
as above. However, one of the cells produced in each divisional cycle receives
almost ALL of the cytoplasm. So, after Meiosis I, there are two haploid cells,
one is very small and the other is very large, having received almost all the
cytoplasm. In fact, the small cell may be so small that it doesn't even have
enough energy to divide again. The big cell DOES complete Meiosis II, but the
cytoplasm is unequally divided again. So, another small cell is produced, and
there is only one large functional egg cell produced. The small, non-functional
cells are called "polar bodies." In many species, like humans, oogenesis stalls
in prophase I. Girls are born with their full complement of egg-producing cells
arrested in prophase I. With the onset of puberty, one cell each month completes
the meiotic cycle and ovulation (eruption of the single egg from the ovary)
occurs. This process of ovulating one egg per month continues throughout a woman's
life until menopause. As such, many cells wait several decades before they complete
meiosis. It is thought that this delay may increase the likelihood of divisional
errors.... the longer a cell waits around, the more likely it is that when it
divides it will not divide correctly. This would explain the increased frequency
of genetic anomalies with increasing maternal age.
E. Sexual Reproduction and Variation
 1.
Meiosis and Mendelian Heredity
1.
Meiosis and Mendelian Heredity
Meiosis was first described in 1876
by Oscar Hertwig; the movement of chromosomes in meiosis was first described
by Eduard Van Beneden in 1888. Sutton and Boveri, working independently in 1902,
saw pairs of chromosomes separating in Meiosis I. Being familiar with the recently
rediscovered work of Gregor Mendel, they appreciated this correlation between
the movement of chromosomes during gamete formation and Mendel's principle of
segregation. If one homolog carried one allele for a trait while the other homolog
carried the other allele, then the separation of homologs could explain the
segregation of alleles during gamete formation. Likewise, if the way that one
pair of homologs separated had no effect on the way that other pairs of homologs
separated, then the movement of chromosomes could explain the principle of independent
assortment, as well.
They hypothesized that these chromosomes,
moving in a pattern consistent and correlated with Mendel's two principles,
might contain the heredity determinants. This is a big deal - it is called the
"Chromosomal theory of heredity". Why do we know that chromosomes
carry the genetic info? Because in 1902, Sutton and Boveri observed the correlation
between the movement of chromosomes and Mendel's laws of heredity, and they
proposed the hypothesis that chromosomes might carry the heredity information
and cause patterns of heredity. This hypothesis was confirmed by
Thomas Hunt Morgan's group working with fruit flies in 1915. Experiments in
1944 determined that it was the nucleic acid in chromosomes, and not the proteins,
that was the hereditary information.
2.
Solving Darwin's Dilemma:
Darwin observed that every population
expressed considerable variation in every generation. He was confused by this,
because if selection had been operating for millennia, why hadn't selection
honed a species to the point where only the best adapted phenotype persisted
and reproduced? In addition, Darwin was a supporter of the idea of blending
heredity. This would further reduce variation each generation (white and black
to grey...). However, because the hereditary material is PARTICULATE and not
a blending fluid, sexual reproduction produces unique combinations of particles
in EVERY generation. Indeed, the amount of new combinations that sexual reproduction
can produce - in other words, the amount of genetic variation that sexual reproduction
can produce - is staggering.
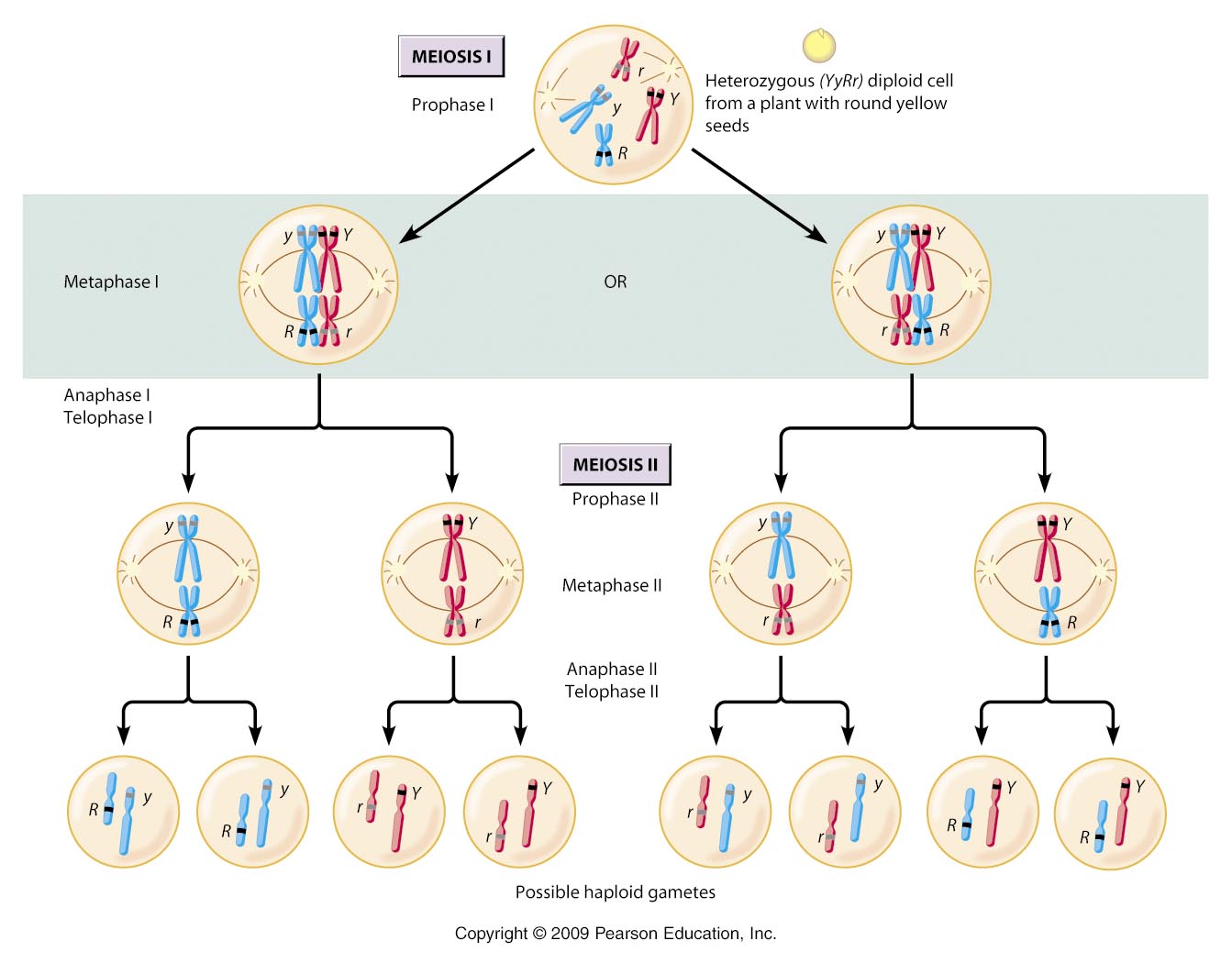
So, a cell 2n = 4, has two pairs of homologus chromosomes. Let's call one pair
'A' and 'a' and the other pair 'B' and 'b'.
***Don't get confused by thinking that these letters represent single genes;
they do not. Rather, they simply represent different chromosomes in a homologous
pair. Because chromosomes carry 100's - 1000's of genes, it is almost impossible
for homologs to be genetically identical over their entire length. So, although
two homologs might carry the same gene at a particular locus (which we could,
confusingly, represent as 'AA'), the homologs will NOT be the same
across all the genes they carry. So, we represent their similarity as homologs
by using the same letter, but their necessary DIFFERENCE with capital and lower
case versions of that letter. OK?***
Now, you know that this organism can produce 4 gametes as a consequence of
segregation and independent assortment: AB, Ab, aB, ab.
A cell that is 2n = 6 has three pairs of homologs, AaBbCc. And, it can produce
8 types of gametes, ABC, abc, Abc, aBC, ABc, abC, AbC, aBc. So, there is a pattern
here. The number of chromosomal combinations that are possible in the gametes
= 2n (two to the 'n' power, where 'n' = the number of chromosomes
in the haploid set). So, for 2n = 4, n = 2, and gamete number = 22
= 4. And, for 2n = 6, n = 3, and gamete number = 23 = 8.
Well, most organisms have lots of chromosomes... humans have 2n = 46. So, n
= 23, and the number of different types of genetic combinations that we EACH
can make in our gametes is 223 = about 8 million. That's more than
the population of Georgia or New Jersey. That is a staggering amount of genetic
variation, produced by each single human being. But of course, one gamete does
not make an offspring; you need two to tango (so to speak). So,
if a male can produce 8 million different types of sperm, and if a female can
produce 8 million different types of eggs, and if any of the sperm are just
as likely to fertilize any of the eggs, then there are 8 million x 8 million
possible combinations of sperm and egg (that's one big Punnett Square!); that's
223 x 223 = 246: about 70 trillion different
types of zygotes that are possible.
That's a LOT of variability, just from one pair of reproducing humans. Now,
the differences in these 8 million gametes are slight. A couple of alleles,
here and there. And, we are still talking about a human genome, so the entire
set of genes still codes for a human; a gene for every trait. So, we are all
fundamentally very similar to one another, sharing 99.9% of our genes. The ways
we differ are actually pretty subtle... the color of hair, the slope of the
nose, the bend of a finger. That's probably why other species seem so homogeneous
to us until we really sit down and try to recognize these little personal differences.
That's the solution to Darwin's Dilemma about the source of variation.... hereditary
particles don't blend together; they remain distinct and form new combinations
from generation to generation, LARGELY AS A CONSEQUENCE OF INDEPENDENT ASSORTMENT,
BUT ALSO AS A CONSEQUENCE OF CROSSING OVER, WHICH WE WILL DESCRIBE LATER.
REALIZE THAT THE ARGUMENTS ABOVE ARE ONLY LOOKING AT THE EFFECTS OF INDEPENDENT
ASSORTMENT. This huge amount of variation is not caused by the production of
a new gene by mutation (although this does occur); this remarkable amount of
variation is produced by recombination: the formation of new combinations of
genes in gametes and offspring during sexual reproduction.
3. Model of Evolution circa 1905 (Rediscovery of Mendel's
Laws)
Source of variation: Independent assortment of particulate hereditary
particles during gamete formation.
Agents of change: NS.
III. Allelic, Genic,
and Environmental Interactions
While Mendel's contributions
were seminal, his postulates and principles did not describe the full range
of complexity that occurs in the patterns of heredity or the expression of genetic
information in the phenotype. Although many genes exhibit simple patterns of
complete dominance, even segregation, and independent assortment, many do not.
In addition, the genes that are received by an organism are not the sole determinant
of the phenotype - even of the characteristics that these genes affect. For
the remainder of this unit, we will look at how Mendelian patterns are modified
(or even violated).
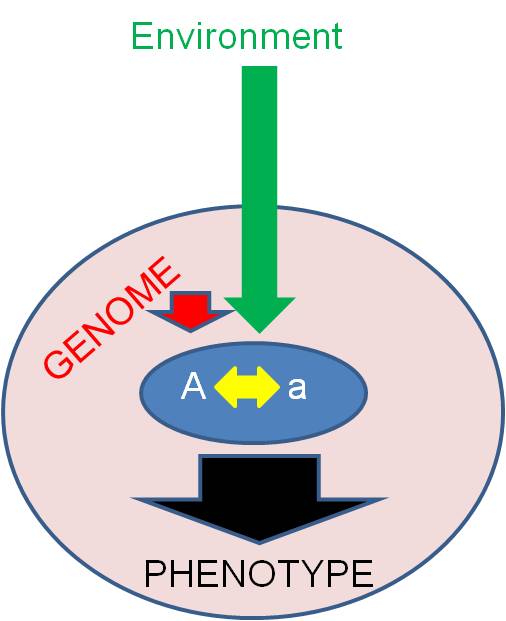 A.
Overview
A.
Overview
Today's lecture will examine some complex
ways in which the genotype is translated into phenotypic expression. The best
way to think about this is to consider a single allele. Now, what we are asking
today is this: what determines whether (and how) this allele is expressed in the
phenotype? The manner in which an allele is expressed in the phenotype is affected
by three broad categories of factors:
- other alleles at the same locus (intralocular interactions)
- other alleles at other loci in the genotype (Interlocular interactions),
- and environmental interactions. These environmental effects may
be proteins in the cytoplasm that were not produced by this genome (maternal/cytoplasmic
effects) or aspects of the extracellular environment.
In addition to considering whether
(and how) an alelle is expressed, we will also consider the "value" of the allele
to the organism (is it 'good' or 'bad'?) This may not be an intrinsic property
of the allele. Rather, the value of an allele is also determined by these three
classes of factors. Keep this in mind; it is the overriding message of transmission
genetics. Unfortunately, our culture has not always appreciated these ideas.
B. Intralocular
Interactions:
Diploid organisms have two alleles
for each locus, or gene. Intralocular effects describe the way that these alleles
interact to cause a phenotype. You are familiar with one type of intralocular
interaction already, so we will start there.
1. Complete
Dominance (Mendelian)
In this case, the heterozygote expresses
a phenotype indistinguishable from the phenotype of the homozygous 'dominant'
individual.
- AA = Aa > aa (Mendel's peas provide examples)
At a cellular level, why does this happen? Well, genes code for proteins. Apparently,
one gene (A - the dominant gene) codes for the production of enough functional
protein for complete cell function and phenotypic expression. Surplus has no
effect. Maybe a reaction is limited by the amount of substrate, and not the
concentration of enzyme (protein). So, one 'dose' of enzymes is enough to metabolize
all of the substrate, and extra enzyme doesn't change this effect. Or the product
of the dominant allele has a higher reactivity with the substrate and always
reacts with it, it's way, even in the heterozygous condition.
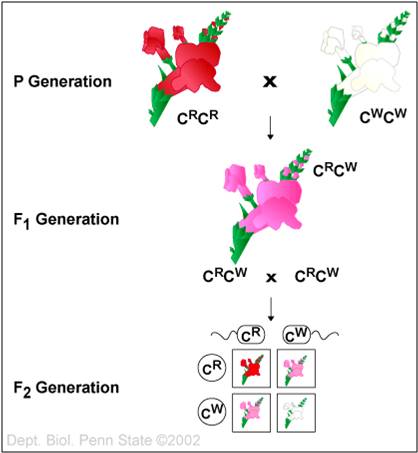 2.
Incomplete Dominance/Intermediate Inheritance
2.
Incomplete Dominance/Intermediate Inheritance
In this case, the heterozygote expresses a phenotype that is intermediate between
the phenotypes expressed by the homozygotes. This is quite common. A classic
example is flower color in 4 o'clocks. Homozygotes for R ("RR") produce
red flowers, homozygotes for "r" (rr) produce white flowers, and heterozygotes
(Rr) produce pink flowers.
- AA > Aa > aa
At a cellular level, this is probably a function of a 'dosage effect'. Two
'on' genes produce more functional product than one, and this surplus influences
cell function and phenotypic expression. Maybe the protein product of an active
R gene is the pigment, itself, and r produces a nonfunctional protein (maybe
it has a premature stop codon and no protein is even produced). So, the two
functional genes in the RR homozygote produce more pigment and a deeper red
color than the single functional gene in the pink heterozygote (Aa).
3. Codominance
In this case, the heterozygote expresses both traits completely; this
does not produce something that is "in between" the homozygotes; rather,
the heterozygote expresses both traits expressed in the homozygotes. The classic
example is the A-B-O blood group in humans. These letters refer to alleles in
the human population that encode a protein that is placed on the surface of
blood cells. These proteins are called 'surface antigens', and they act as a
chemical signal to white blood cells that these cells are "self".
There are three alleles in the human population at this locus. Of course, each
diploid person only has two of these three alleles. The genotypes that are possible,
and the phenotypes they express, are as shown:
| Genotype |
Interaction between alleles |
Phenotype |
| AA |
|
A |
| AO |
A dominant to O |
A |
| BB |
|
B |
| BO |
B dominant to O |
B |
| OO |
|
O |
| AB |
A and B codominant |
AB |
The A allele codes for the A surface antigen. The B allele codes for the B
surface antigen. The O allele is non-functional; no surface antigens are produced.
So, any genotype with an A allele makes A surface antigen; any genotype with
a B makes B surface antigen. A genotype that is AB has different active genes,
each producing their own surface antigen, and so BOTH A and B surface antigens
are affixed to the outside of the cell. It is a phenotype that is BOTH A and
B at the same time.
A more general cellular explanation is this. The allelic products are both
functional and work on slightly different substrates. Having both gives the
phenotype a qualitative diffence, not a quantitative difference. This is often
confused with "incomplete dominance", but think about it this way:
"pink" flowers are neither "red" nor "white" -
they are something different that isn't red or white. But AB blood is BOTH A
and B at the same time, not different from A and B, but a combination of both
A and B.
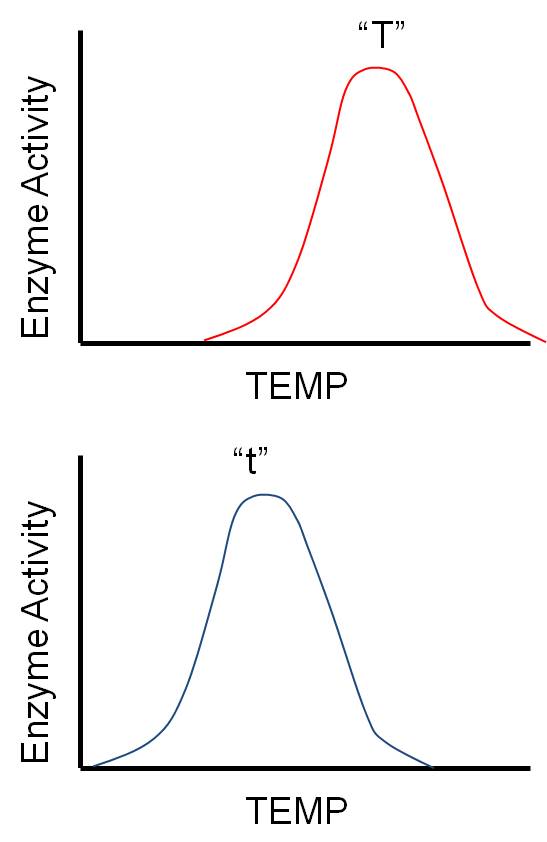 4.
Overdominance (Heterosis)
4.
Overdominance (Heterosis)
In this seemingly unusual situation, the heterozygote expresses a phenotype
"more extreme" than either homozygote. So, if AA = tall and aa = short, then
the heterozygote Aa = Tallest.
This may seem unusual, but it is actually easy to explain. Suppose the two
alleles work slightly differently, or under slightly different conditions. For
instance, consider a gene that codes for an enzyme that influences growth. As
you can imagine, there are LOTS of such genes! Almost any catalyst that affects
metabolism, respiration or protein synthesis will influence growth. Maybe
the two alleles for this enzyme work at different temperatures (H = warm and
h = cold). If the organism lives in an environment that is warm most of
the time, then the homozygote for HH will grow most of the time, and will be
taller than the hh homozygotes that only grow during the rare colder periods.
However, the heterozygote Hh grows ALL the time, when it is cold and warm, and
so is taller than either homozygote. Variation at a locus can be as advantageous
as variation in offspring; variation in the genotype (heterozygosity) may allow
the organism to cope with a greater range of environmental conditions, or metabolize
more substrates. Thus, there is often a 'heterozygote advantage' at certain
loci.
B. Interlocular
Interactions:
The effect of an allele can be influenced
by other genes in the genome, at other loci. These are called "inter"-locular
effects.
1. Quantitative
"polygenic" effects
There are many proteins (and RNA's)
that are valuable in high concentrations; higher concentrations than production
from a single locus can produce. Selection has favored organisms in which these
genes have been duplicated (through a process we will examine later). The organisms,
with two or several different loci that produce the same protein (or RNA), can
produce LOTS of that essential molecule. A good example is r-RNA. The production
of all proteins in the cell depends on ribosomes, and they all carry r-RNA. So,
selection has strongly favored organisms that have multiple copies of the genes
that encode r-RNA; they can produce LOTS of r-RNA, and thus produce LOTS of ribosomes...
and this increases the rate that they can produce all of their proteins. Now,
for genes that encode proteins that influence a phenotypic characteristic, multiple
genes means that more than one locus produces the protein product and influences
that trait. So, the final phenotype is determined by the cumulative, "quantitative"
sum of all the genes acting on this trait.
As a consequence of many loci acting on a trait, there are many more combinations
that are possible - resulting in a wide variety of phenotypic expression that
forms a nearly 'continuous' range of variation.
For example, consider human skin pigmentation. There are at least 16 loci that
code for melanin (skin pigment) production. Although there is complete
dominance at each locus, this multiplication of genes allows for an extraordinary
and continuous amount of phenotypic variation, from all 16 genes 'on' (and very
very dark skin), to 15 'on' and 1 'off' ( very dark skin), to 14 'on' and 2
'off' (a bit lighter), to , 13:3, etc. , all the way to 0 'on' and 16 'off'
(no pigments produced at all).
Humans evolved on the plains of Africa.
The loss of hair was adaptive (to decrease insulation in the subtropical heat),
but this made the skin vulnerable to UV rays that cause mutation. Selection
favored humans that could make lots of melanin, to protect the skin against
these UV rays. Selection favored humans that duplicated their melanin genes,
and could thus produce more pigment. Humans that continued to live in tropical
areas continued to benefit from this dark skin. In cooler climes with less intense
sun, selection favored humans that spent less energy on melanin production,
selecting for the recessive alleles at each of these multiple loci. In addition,
lighter skinned people benefitted at these latitudes by using the sun's energy
to synthesize vitamin D in the skin.
2. Epistasis
 In
epistasis, one locus has precedence over the expression at another locus and
can override it. Albinism is the classic example. Albinism ooccurs in all populations
of humans (and many other animals). It usually does NOT involve the melanin
producing genes. (So, true albinism is NOT caused by the 0 'on', 16 'off' scenario
described above). Rather, it involves another locus. An 'aa' individual at the
albinism locus does not make the precursor for melanin. So, with no
precursor from which melanin can be made, it doesn't matter WHAT the genotypes
are at the 16 melanin loci - there won't be any melanin produced. This is why
albinism occurs in all human populations (or 'races') regardless of of the skin
color typical for that population - because it is influenced by a gene that
is inherited independent of the melanin producing genes.
In
epistasis, one locus has precedence over the expression at another locus and
can override it. Albinism is the classic example. Albinism ooccurs in all populations
of humans (and many other animals). It usually does NOT involve the melanin
producing genes. (So, true albinism is NOT caused by the 0 'on', 16 'off' scenario
described above). Rather, it involves another locus. An 'aa' individual at the
albinism locus does not make the precursor for melanin. So, with no
precursor from which melanin can be made, it doesn't matter WHAT the genotypes
are at the 16 melanin loci - there won't be any melanin produced. This is why
albinism occurs in all human populations (or 'races') regardless of of the skin
color typical for that population - because it is influenced by a gene that
is inherited independent of the melanin producing genes.
Multiple loci may influence genes
in interactive ways, not just in quantitative, additive ways
like skin color. For example, in sweet peas (not the garne peas of Mendel),
there are two genes that influence flower color. Both exhibit complete
dominance, and both proteins must be produced in order for purple flower color
to be expressed. So:
aaBB (white) x AAbb (white)
|
100% AaBb (purple)
|
9/16 A_B_ (purple)
7/16 aaB_ or A_bb or aabb (white)
(In these examples, the underlined space means that it
doesn't matter what the second allele is... if one allele at a locus is dominant,
then in a completely dominant system, you will get the same phenotype regardless
of whether the second allele is dominant or recessive. So, rather than writing
all the separate genotypes out that yield the same phenotype, you just put the
genes that determine the phenotype. Of course, to express the recessive phenotype,
you have to be homozygous - so to represent genotypes that express the recessive
phenotype, you need to write both alleles. aaB_ refers to aaBB and aaBb... they
both yield the aB phenotype.)
 C.
Environmental Interactions
C.
Environmental Interactions
The environment has many direct effects
on the phenotype. A fox may change from white to brown because a bucket of white
paint falls on it's head. That would be a direct environmental effect; it happened
independently of anything going on with the genotype. Those are NOT the effects
we are talking about here. Rather, we are talking here about an interactive
effect between the environment and the genes at a locus, such that the environment
changes how the genotype is expressed as the phenotype.
1. Temperature
In arctic fox, the brown summer fur
turns to white as temperatures drop. This is NOT a direct effect of temperature
change on pigment in the hair shaft. Rather, the change in temperature changes
how the genes and their protein products work. As temperatures drop, melanin
genes are turned off, or the enzymes that catalyze the production of melanin
change shape and no longer function. The result is that no melanin is produced,
and thus the fur turns white.
2. Toxins
Susceptibility to certain mutagens
will vary with the genotype. So, two people who are homozygous for a type
of lung cancer (and 'should' genetically express that cancer) might have very
different phenotypes. One, who smokes and exposes their lungs to mutagenic
compounds might trigger those cancer genes. The other person, however,
with the same genotype, might not have cancer because they never smoked and
thus never exposed themselves to the environmental "trigger". Likewise,
someone homozygous for an alternative allele might smoke without developing
lung cancer.
These effects
may be "probabilistic". For instance, some genes associated with cancers
are only expressed (cause cancer) in a fraction of the individuals with that
genotype. This could be due to different environmental exposure, but it
could also be caused by different genes at other loci that interact with this
cancer gene and augment or supress its effects through interlocular interactions.
D.
The 'Value' of an Allele
1. Lethal
Alleles: Some alleles are ALWAYS bad.
Usually, these have rather profound
effects on basic cellular metabolism, affecting a vital enzyme or structural
protein. Typically, lethal alleles that are DOMINANT get weeded out of the population
very quickly by selection (anyone that gets one allele dies...). So, most lethal
alleles that are maintained in natural populations are recessive, and thus are
"hidden" in heterozygotes and are be passed down through generations.
However, some lethal alleles ARE dominant. Can you think of another way that
an allele could 'escape detection' by selection (which is "differential
REPRODUCTIVE success"?). Huntington's Chorea is a dominant lethal neurodegenerative
condition. Actually the Homozygote dies in utero. However the heterozygotes
survive and are phenotypically normal into their 50's. THEN, only AFTER they
might have reproduced and passed the gene on, do the effects of this dominant
gene start to come forward. So, they are "invisible" to selection
because they are expressed POST-REPRODUCTIVELY.
2. Environmental Effects:
The value of an allele often depends on the environment
In Sickle Cell Anemia, red blood
cells 'sickle' when the concentration of oxygen in the bloodstream drops.
This occurs when the person is active and the oxygen demand by muscle increases,
drawing oxygen out of the bloodstream and depressing oxygen concentration in
the cells in the bloodstream. The
sickle-shaped cells do not pass through capillary beds as easily as normal red
blood cells; they clog capillaries, resulting in oxygen deprivation and tissue
damage downstream from the clot. This is usually most pronounced in the
liver, kidneys, and brain, and eventually often results in premature death.
Sickle cell anemia is caused by an
altered beta globin allele, which causes a single amino acid change in the beta-globin
proteins in hemoglobin. The trait exhibits incomplete dominance - one
"s" allele will result in some "sickling" of red blood cells at low oxygen concentration,
but the condition is not nearly as severe as it is in the homozygous condition.
Now, you might think that even one
sickle-cell allele would 'always be bad' - after all, I just said that heterozygotes
do suffer some debilitating effects. But in this case, the "value"
of a sickle cell allele - whether it is 'good' or 'bad' - depends on the environment.
In particular, it depends on the presence of the Plasmodium parasite
that causes malaria.
In the tropics, a primary source
of human mortality is malaria. In 2006, nearly 900,000 people died of
malaria; over 91% of these deaths were children in Africa. There were an estimated
250 million cases reported globally. Malaria is caused by several species
of protists in the genus Plasmodium. This single-celled parasite
is transmitted by female mosquitos. When they bite a human to take a blood meal
(only female mosquitos drink blood - they use the protein to nourish their developing
eggs), the parasites enter the human host's bloodstream and infects red blood
cells. They divide mitotically, producing hundreds of offspring and eventually
rupturing the cell. Thus, infection causes extreme loss of RBC's - "anemia"
(don't get this confused with sickle cell anemia; we are just talking about
malaria right now!!). Curiously, the Plasmodium parasite can
not reproduce in cells with the altered form of "sickle-cell" hemoglobin - even
in the heterozygous condition. So, the heterozygote suffers some sickling
on occasion, but is protected from malaria. In Africa, SS homozygotes
have lower survivorship than the Ss heterozygotes because the SS individuals
are exposed to malaria. The ss homozygotes have lower survivorship than
the Ss heterozygotes because of the more pronounced debilitating effects of
the sickle cell disease.
So, is an 's'
allele "good" or "bad"? Well, that depends on other alleles at that
locus (if it's with another 's' allele it is always bad), but it also depends
on the environment. If the 's' is with an 'S' in the temperate zone, it
is bad (relative to the reproductive succes of the normal SS homozygote)....
but if the 's' allele is paired with an 'S' allele in the tropics, then it is
"good" - better than having two dominant alleles (SS).
TEMPERATE ZONE: survivorship: SS > Ss > ss
TROPICS: survivorship: SS < Ss > ss
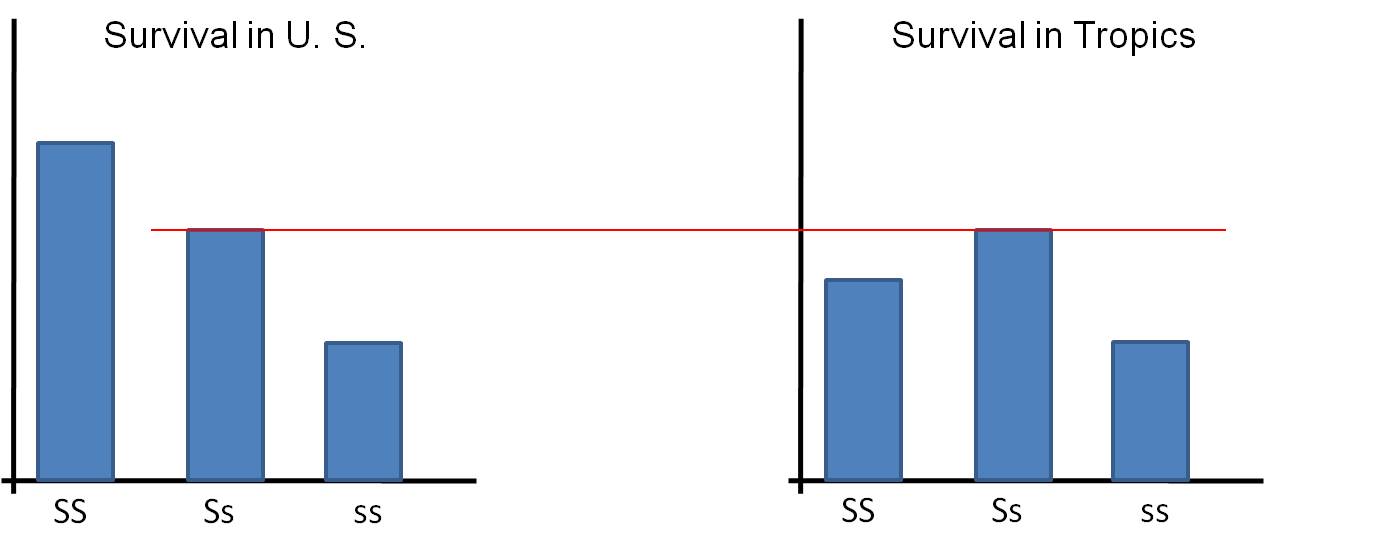
You should relate this to the corollary of Darwin's Theory of Natural Selection.
Populations in different environments will diverge from one another genetically,
as the environment selects for different traits (genotypes).
E. Summary:
An organism is more than just the
sum of their parts, even at a genetic level. How the genotype works is not just
the additive sum of all the genes acting independently. Rather, the way some
genes work depends on the other genes in the genotype. The phenotype - what
a complex organism IS - is the result of these complex interactions between
genes, with the additional layer of environmental interactions and direct environmental
effects. Indeed, given the potential complexities involved in how 1000's of
genes and a complex environment can interact, it is rather surprizing that many
genetic effects are simple enough to model as independent entities with a Punnet
Square - without considering the other genes or the environment. So, Mendelian
Genetics are actually the easiest, simplest patterns in heredity to recognize:
and that is probably why they were recognized first.
Study Questions.
1) Explain how a continuously
variable trait could be governed by genes.
2) What is
an epistatic interaction? Give an example.
3)
How can the environment influence the expression of a trait?
4) How can
the environment influence the VALUE of a trait? Relate this to Darwin's
idea of the diverge of populations in different environments.
5) Why are
most lethal alleles recessive? Answer with respect to the effects of selection
on a dominant, deleterious gene.
6) As such,
how can a dominant lethal allele be maintained in a population?
7)
Consider
this cross:
AaBbCc x AaBbCC
- assume independent assortment of the three genes
- There is incomplete dominance at the A locus (meaning A is incompletely
dominant to a).
- There is complete dominance at the B locus.
- There is overdominance at the C locus.
How many genotypes are possible
in the offspring?
How many phenotypes are possible
in the offspring?
8) Conduct
the following cross: Aa x Aa
Determine the genotypic and phenotypic
ratios if there is:
- complete dominance
- Incomplete dominance
- Codominance
- Overdominance
9) Provide
a cellular explanation for overdominance.
 2.
Meiosis I (Reduction)
2.
Meiosis I (Reduction)  4.
Meiosis II: (Division):
4.
Meiosis II: (Division):  a.
Spermatogenesis:
a.
Spermatogenesis:
 1.
Meiosis and Mendelian Heredity
1.
Meiosis and Mendelian Heredity A.
Overview
A.
Overview 2.
Incomplete Dominance/Intermediate Inheritance
2.
Incomplete Dominance/Intermediate Inheritance
 4.
Overdominance (Heterosis)
4.
Overdominance (Heterosis)
 In
epistasis, one locus has precedence over the expression at another locus and
can override it. Albinism is the classic example. Albinism ooccurs in all populations
of humans (and many other animals). It usually does NOT involve the melanin
producing genes. (So, true albinism is NOT caused by the 0 'on', 16 'off' scenario
described above). Rather, it involves another locus. An 'aa' individual at the
albinism locus does not make the precursor for melanin. So, with no
precursor from which melanin can be made, it doesn't matter WHAT the genotypes
are at the 16 melanin loci - there won't be any melanin produced. This is why
albinism occurs in all human populations (or 'races') regardless of of the skin
color typical for that population - because it is influenced by a gene that
is inherited independent of the melanin producing genes.
In
epistasis, one locus has precedence over the expression at another locus and
can override it. Albinism is the classic example. Albinism ooccurs in all populations
of humans (and many other animals). It usually does NOT involve the melanin
producing genes. (So, true albinism is NOT caused by the 0 'on', 16 'off' scenario
described above). Rather, it involves another locus. An 'aa' individual at the
albinism locus does not make the precursor for melanin. So, with no
precursor from which melanin can be made, it doesn't matter WHAT the genotypes
are at the 16 melanin loci - there won't be any melanin produced. This is why
albinism occurs in all human populations (or 'races') regardless of of the skin
color typical for that population - because it is influenced by a gene that
is inherited independent of the melanin producing genes.
 C.
Environmental Interactions
C.
Environmental Interactions