Modern Evolution I:Population Genetics
I. Modern Evolution: Population Genetics
A. Overview
As a consequence of our modern understanding
of heredity and genetics, we have learned quite a bit about variation AND evolution,
and our model, a this point in the class is:
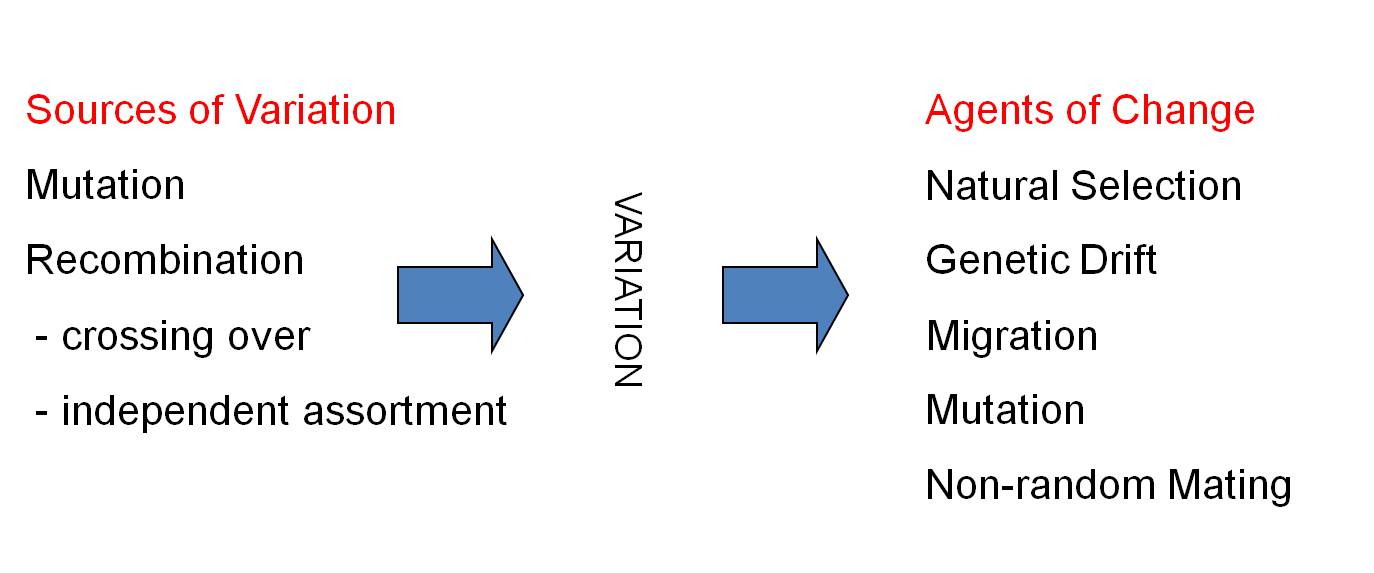
Sources of Variation Agents of Change
MUTATION:
-New Genes:
Natural Selection
point mutation
Mutation (polyploidy can make new species)
RECOMBINATION:
- New Genes:
exon shuffling
-New Genotypes:
-crossing over
- independent assortment
 In
the early 20th century, at the same time that T. H. Morgan was studying mutations
and creating linkage maps, other biologists were considering the evolutionary
implications of this new knowledge regarding genetic variation. They appreciated
that individuals do not evolve - evolution is a process that occurs at the population
level. For example, as a consequence of differential reproductive success among
individuals in a population, the range of phenotypes and their relative frequencies
in the population will change over time. Individuals are born, life, reproduce
(maybe) and die. As a result of passing on their genes at different frequencies,
the genetic structure of the population changes over time (evolution). Two biologists,
G. Hardy and W. Weinberg, constructed a model to explain how the genetic structure
of a population might change over time.
In
the early 20th century, at the same time that T. H. Morgan was studying mutations
and creating linkage maps, other biologists were considering the evolutionary
implications of this new knowledge regarding genetic variation. They appreciated
that individuals do not evolve - evolution is a process that occurs at the population
level. For example, as a consequence of differential reproductive success among
individuals in a population, the range of phenotypes and their relative frequencies
in the population will change over time. Individuals are born, life, reproduce
(maybe) and die. As a result of passing on their genes at different frequencies,
the genetic structure of the population changes over time (evolution). Two biologists,
G. Hardy and W. Weinberg, constructed a model to explain how the genetic structure
of a population might change over time.
Their model begins by constructing
an 'equilibrium' model - a model of what the gentic structure would look like,
and how it would behave, if there was NO CHANGE over time. (We can liken this
to a "stitstical null hypothesis of no effect"). Then, an actual population
is compared to this model, so see whether the population is evolving or not.
B. The Genetic Structure of a Population
Our first step is to describe the
genetic structure of a population; we need to do this before we can model what
it would do over time. The genetic structure of a population is defined by the
gene array and the genotypic array. To understand what these are, some definitions
are necessary:
1. Definitions:
- Evolution: a change in the genetic structure of a population
- Population: a group of interbreeding organisms that share
a common gene pool; spatiotemporally and genetically defined
- Gene Pool: sum total of alleles held by individuals in a population
- Genetic structure: Gene array and Genotypic array
- Gene/Allele Frequency: % of alleles at a locus of a particular
type
- Gene Array: % of all alleles at a locus: must sum to 1.
- Genotypic Frequency: % of individuals with a particular genotype
- Genotypic Array: % of all genotypes for loci considered;
must = 1.
2. Basic
Computations - Determining
the Genotypic and Gene Arrays:
 The
easiest way to understand what these definitions represent is to work a problem
showing how they are computed.
The
easiest way to understand what these definitions represent is to work a problem
showing how they are computed.
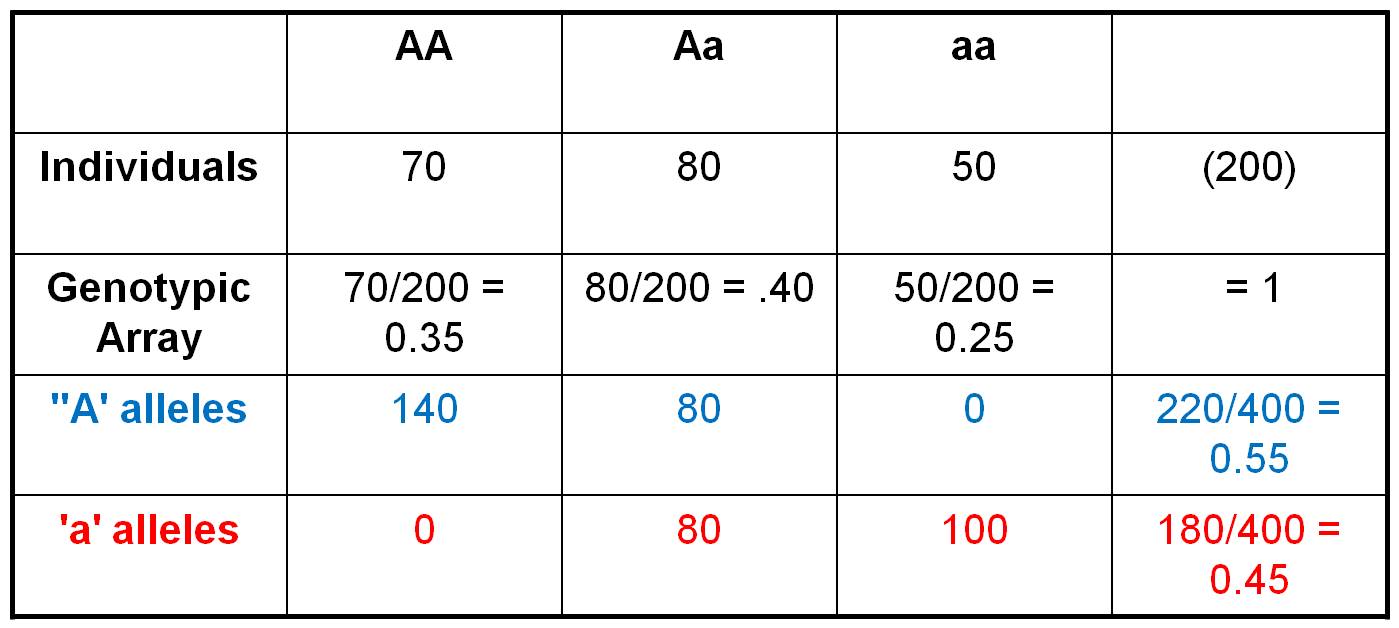
Consider the population shown to
the right, in which there are 70 AA individuals, 80 heterozygotes, and 50 aa
individuals. We can easily calculate the Genotypic Frequencies by dividing each of these values by the total number of individuals in the population.
So, the Genotypic Frequency of AA = 70/200 = 0.35. If we account
for all individuals in the population (and haven't made any careless math erros),
then the three genotypic frequencies should sum to 1.0. The Genotypic
Array would list all three genotypic frequencies: f(AA) = 0.35,
f(Aa) = 0.40, f(aa) = 0.25. A Gene Frequency is the
% of all genes in a population of a given type. This can be calculated two ways.
First, let's do it the most obvious and direct way, by counting the alleles
carried by each individual. So, there are 70 AA individuals. Each carries 2
'A' alleles, so collectively they are 'carrying' 140 'A' alleles. The 80 heterozygotes
are each carrying 1 'A' allele. And of course, the 'aa' individuals aren't carrying
any 'A' alleles. So, in total, there are 220 'A' alleles in the population.
With 200 diploid individuals, there are a total of 400 alleles at this locus.
So, the gene frequency of the 'A' gene = f(A) = 220/400 = 0.55.
We can calculate the frequency of the 'a' alleles the same way. The 50 'aa'
individuals are carrying 2 'a' alleles each, for a total of 100 'a' alleles.
The 80 heterozygotes are each carrying an 'a' allele, and the 140 AA homozygotes
aren't carrying any 'a' alleles. So, in total, there are 180 'a' alleles out
of a total of 400, for a gene frequency f(a) = 180/400 = 0.45. The gene
array presents all the gene frequencies, as: f(A) = 0.55, f(a)
= 0.45.
There is a faster way to calculate
the gen frequencies in a population than adding up the genes contributed by
each genotype. Rather, you can use these handy formulae:
f(A) = f(AA) + f(Aa)/2
f(a) = f(aa) + f(Aa)/2
So, to calculate the frequency of
a gene in a population, you add the frrequency of homozygotes for that allele
with 1/2 the frequency of heterozygotes. In our example, this would be:
f(A) = 0.35 + 0.4/2 = 0.35 + 0.2
= 0.55
f(a) = 0.25 + 0.4/2 = 0.25 + 0.2
= 0.45
Wow... that's alot faster.
C. The Hardy-Weinberg
Equilibrium Model
1. Goal:
The goal of the "Hardy-Weinberg
Equilibrium Model" (HWE) is to describe what the genetic structure of the
population would be if NO evolutionary change occurs. Working independently, Hardy and Weinberg realized
that the gene frequencies in a population will NOT change - will remain in EQUILIBRIUM
- if the following conditions are met:
- there is random mating
- no selection
- no mutation
- no migration
- and the population is infinitely
large.
And, they realized that a population
will reach an equilibrium in GENOTYPIC frequencies, too, after one generation
of meeting these expectations (and for as long as it does so), a population
will NOT EVOLVE. Let's see how they came by these conditions.
 2.
Example:
2.
Example:
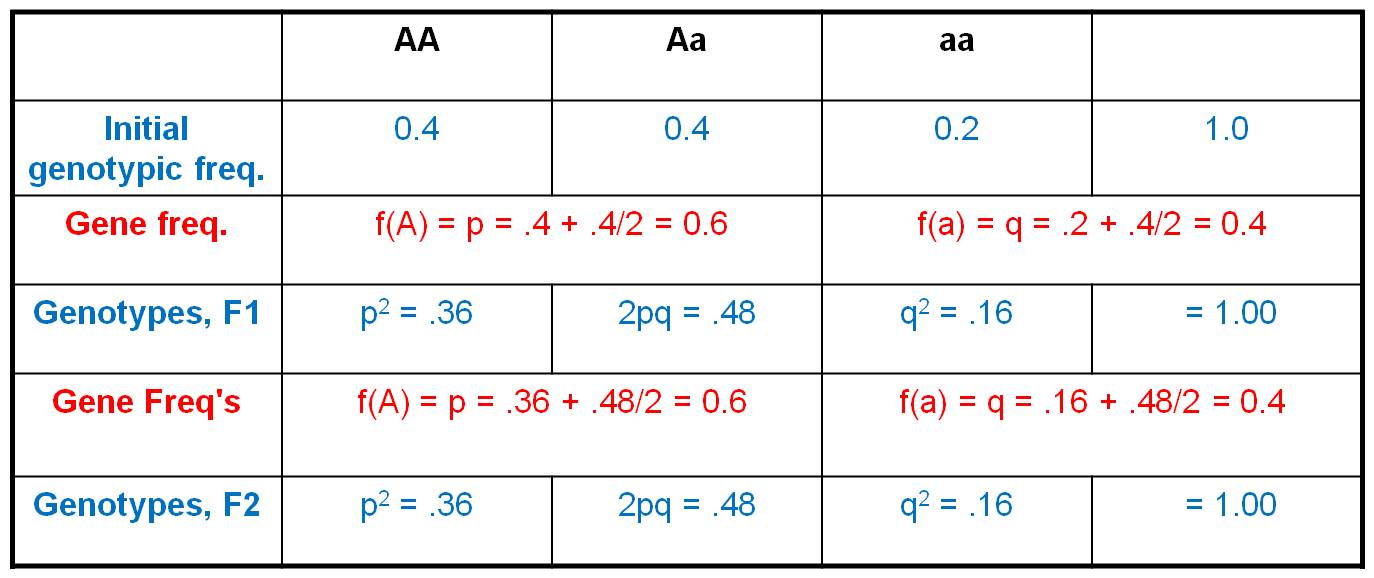
Consider the initial population,
with a genotypic array as shown. The gene frequencies are:
A = 0.4 + (0.4/2) = 0.6
a = 0.2 + (0.4/2) = 0.4
Now, consider this gene pool in which
60% of the alleles are 'A' and 40% of the alleles are 'a' (as defined by the
gene frequencies).
So, now we employ the HWE model.
IF the population mates at random, then we can use the product rule to determine
the probability of any two gametes coming together. The propability that and
'A' sperm fertilizes an 'A' egg = 0.6 x 0.6 = 0.36. And of course, this is the
only way to produce an 'AA' zygote. So, the frequency of 'Aa' zygotes (the F1
offspring) produced by this population should be 0.36. Likewise, the probability
that an 'a' sperm fertilizes an 'a' egg = 0.4 x 0.4 = 0.16. And again, this
is the only way to make an 'aa' zygote, so the total frequency of 'aa' zygotes
in the F1 will be 0.16. Now, there are two ways to make an 'Aa' zygote: an 'A'
sperm can fertilize an 'a' egg (probability = 0.6 x 0.4 = 0.24), and an 'a'
sperm can fertilize an 'A' egg (also with a probability of 0.4 x 0.6 = 0.24).
So, the total frequency of Aa zygotes in the F1 will be 2 x 0.24 = 0.48. If
we generalize, and let f(A) = p and f(a) = q, then the genotypic frequencies
under HWE can be calculated as: f(AA) = p2, f(Aa) = 2pq, and f(aa)
= q2.
Now, of course, these calculations
will only be true IF the population mates at random. AND, they will only be
true if there is no mutation. If 'A" alleles are mutating into 'a' alleles,
then the gene frequencies will not be 0.6 and 0.4, and calculations based on
these numbers will not be correct. So, we must assume NO MUTATION. Likewise,
we can't have any migration; we can't have 1000 AA individuals migrate into
our population, or that would change the gene frequencies, too; and our predictions
based on frequencies of 0.6 and 0.4 would be incorrect. So, we must assume NO
MIGRATION, too.
So, at this point we have zygotes
at the frequencies shown in the "Genotypes, F1" row. In order for
there to be no change in the genetic structure of the population, there must
be NO SELECTION. In other words, all genotypes must have the same probability
of survival and reproduction. Only then will they contribute gametes at frequencies
of p = 0.6 and q = 0.4. (If there were selection, and if AA individuals were
the only zygotes to survive to reproeuce, for instance, then the gene frequencies
would change and our predictions based on frequencies of 0.6 and 0.4 would not
be correct).
And finally, this model will only
be explicitly true for populations that are infinitely large: because that is
the only time when we can be garaunteed that predictions based on random chance
will be exactly met. (Think about it this way... suppose I give you a coin that
is absolutely perfectly balanced at the molecular level. It IS PERFECTLY BALANCED.
And suppose I ask you, "how many times do you have to flip that coin to
be ABSOLUTELY SURE of producing a 50:50 ratio of heads to tails? Well, if you
only flip it four times, you know that, just by chance, you would often get
3 heads and a tail or 3 tails and a head. And even if you flip it 10,000 times,
you might get 5001 heads and 4999 tails, even though the coin is perfectly balanced.
To be absolutely garaunteed that the predictions of this probabilisitic model
will be met exactly, you must flip the coin an infinite number of times. Obviously,
this is a theoretical constraint because no population is infinitely large.
But this is a theoretical model of no change, so we can employ theoretical expectations.
The same is true of our 'expectation' of a perfectly balanced coin - this expectation
will only be met, for sure, in an infinitely large sample. Yet we continually
employ that expectation for a perfectly balanced coin.
So, that is why these assumptions
exist. It is only when these are met that a population will not evolve. Wow.
That should seem rather amazing. It is only when these assumptions are ALL met
that a population WON'T change. If any of these assumptions is not met, a population's
genetic structure WILL change... and that is evolution. So, from this analysis,
we should expect populations to evolve - it is only under a rare combination
of events (no, mutation, no selection, no migration, random mating, and an infinitiely
large population) that evolution WON'T happen.
3. Utility
- If no real populations can explicitly
meet these assumptions, how can the model be useful? For instance, no
real population is infinitely large, so how can the model be useful? We
use it for COMPARISON. This model describes what the genotypic frequencies
should be IF the population was in equilibrium.
If the real genotypic frequencies are not close to these expectations, then
the population is not in HWE.... it is evolving. And if a population is not
in HWE, then the population must be violating one of the assumptions of the
HWE model. Think about that. The HWE is only 'true' if the assumptions
are being met. If your real population differs from the model, then one
of the assumptions must not apply to your real population. This narrows
your focus on WHY the real populations isn't behaving randomly... and it might
identify WHY the population is evolving.... which is a biologically interesting
question.
 -
Again, the coin analogy applies. No REAL coin is probably exactly perfectly
balanced. But, if I give you a coin and ask you how balanced it is, you flip
it a few times and compare its behavior to WHAT YOU WOULD EXPECT FROM A PERFECT
COIN (50:50 RATIO). Even though a perfectly balanced coin does not exist, we
can use this theoretical model as a benchmark, to compare the behavior of real
coins. Many real coins act in a manner that is consistent enough with
the expectations from a perfectly balanced coin that we are willing to use them
AS IF they were perfectly balanced. The Hardy Weinberg Equilibrium
Model is the same... it is a model of no change against which we can measure
real populations.
-
Again, the coin analogy applies. No REAL coin is probably exactly perfectly
balanced. But, if I give you a coin and ask you how balanced it is, you flip
it a few times and compare its behavior to WHAT YOU WOULD EXPECT FROM A PERFECT
COIN (50:50 RATIO). Even though a perfectly balanced coin does not exist, we
can use this theoretical model as a benchmark, to compare the behavior of real
coins. Many real coins act in a manner that is consistent enough with
the expectations from a perfectly balanced coin that we are willing to use them
AS IF they were perfectly balanced. The Hardy Weinberg Equilibrium
Model is the same... it is a model of no change against which we can measure
real populations.
If HWE can be assumed, then the frequency
of recessive diseases can be assumed to equal q2, and the frequency of carriers
in the population can be estimated like this:
1) The frequency of hemachromatosis
worldwide is 1/450. If we assume that hemochromatosis is caused by a recessive
gene (q), and if we assume the population is in HWE with respect to this trait,
then q2 = 1/450 = 0.002. So, we take the square-root of both sides
to find q = 0.047. Well, if q = 0.047, and if p + q = 1, then p = 1 - 0.047
= 0.953.
2) If q = 0.047 and p = 0.953, then
the frequency of heterozygous carriers = 2pq = 0.09. So, we estimate that 9%
of the population are carriers.
Now, you might say, "but we
just determined that HWE would be unusual; so why would we assume it is
true for a given gene?" Well, a deleterious gene has already been
largely weeded out of a population, so selection against the few alleles that
are left is really weak. Indeed, this condition may not influence reproductive
success, anyway (NO SELECTION). In addition, we don't select mates based on
whether they have hemochromatosis (I bet you NEVER asked your date if they have
hemochromatosis, for example!!), so we can assume there is RANDOM MATING in
the population with respect to this trait. And although the human population
is not infinite, it is really big (6.9 BILLION), so the effect of sampling error
is probably very small. Mutation is very rare, so the effects of mutation are
likely to be very small. And if we are making an estimate based on the whole
human population, then there can be no 'migrants' coming in from somewhere else
(Martians?). So, in some cases, we can reasonably assume a population might
be in HWE for a given gene. Of course, we could be wrong... and we would test
that prediction by sampling individuals in the population and determining the
frequency of heterozygotes genetically. But at least we would have a working
hypothesis.
D. Deviations
From HWE:
1. Mutation
Although large scale mutations like
polyploidy can cause instantaneous speciation, what we are talking about here
are substitution mutations that change one allele into another, or make a new
allele. Although such changes are very important
sources of new variation, they do not change the genetic structure of a population
very much at all, even when they occur: these mutations are rare, usually occurring
at a rate of 1 x 10-4 to 1 x 10-6.
Consider a population with:
f(A) = p = 0.6
f(a) = q = 0.4
Suppose 'a' mutates to 'A'
at a realistic rate of: µ = 1 x 10-5 . How will this rate of
mutation change gene frequencies? Not much: 'a' will decline by: qm = .4 x 0.00001
= 0.000004
'A' will increase by the same amount. So, the new gene frequencies will be:
p1 = p + µq = .600004, and q1 = q - µq = q(1-µ) = .399996.
So, mutation is a very important source of new alleles, but it doesn't change
the gene frequencies in a population very much.
2. Migration
Consider a resident population in
which p = 0.6 and q = 0.4. Suppose immigrants migrate into this population,
bringing A and a alleles into the population at these frequencies: p =0.8 and
q = 0.2. The effect of this influx will depend on the number of immigrants relative
to the number of residents. 100 immigrants may not change the genetic structure
of a population containing 1 million residents, but they could have a dramatic
effect on a population of 100 residents. We measure this relative effect by
quantifying the proportion of the total combined population that are immigrants.
So, in our example, suppose so many immigrants move in that they represent
10% of the new, combined population. We calculate new p as a weighted
average based on fraction of immigrants and residents:
So, p1 = (0.6)(0.9) + (0.8)(0.1)
= 0.54 + 0.08 = 0.62
residents contribute p at a rate of 0.6, and they represent 90% of the combined
population. Immigrants contribute p at a rate of 0.8, and they are 10%
of the population.
q1 = (0.4)(0.9) + (0.2)(0.1)
= 0.36 + 0.02 = 0.38, so we have done our math right because 0.62 + 0.38 = 1.0
There are two possible evolutionary
effects. First, migration will make two populations similar to one another;
particularly if the rate of immigration is high or the process is continuous
over time. Migration can also introduce new alleles into a population, but again
this effect will be correlated with the abundance of immigrants relative to
the number of residents.
 3.
Non-Random Mating:
3.
Non-Random Mating:
a. Positive Assortative Mating
There are many ways that non-random
mating can occur. We will look at a couple. The first example is called "positive
assortative mating". This is where mates 'sort' themselves with others
of the same genotype. This can be thought of as "like mates with like".
So, consider our old four o'clock plants with incomplete dominance. A population
might contain red, pink, and white flowers. Suppose the red flowers open in
morning, and are pollinated just by hummingbirds (that prefer red flowers).
Suppose the white flowers open at night, and are pollinated by moths. And suppose
the pink flowers open in the afternoon, and are pollinated by bees and butterflies.
In this case, "like mates with like" for flower color (and time of
opening). So, a plant with red flowers will only mate with another plant having
the same genotype for (red) flower color. Now, it is IMPORTANT to realize that
plants are only positively assorting for flower color and opening time in this
case. One red flowering plant may be tall while the other is short; one may
have hairy leaves while the other has smooth leaves. Indeed, the plants may
be mating at random with respect to all other traits.
When AA individuals mate only with
each other, all their offspring will be AA, as well. So, if 20% of the population
is AA (intial genotypic frequency = 0.2), and if there is no difference in reproductive
success (because we are only violating the assumption of random mating so there
is no selection), then these parents will make 20% of the offspring and they
will all be AA. The same goes for aa individuals only mating with other aa individuals
- all their offspring are aa. However, when Aa heterozygotes only mate with
one another, they produce AA, Aa, and aa offspring in a 1/4:1/2:1/4 ratio. If
60% of the population is heterozygous, then they will make 60% of the offspring...
but these offspring won't all be heterozygous; only 1/2 - or 30% will be heterozygous.
15% will be AA and 15% will be aa. So, the total frequency of AA offspring in
the F1 will be 35%; 20% had AA parents and 15% had Aa parents.
As a consequence of positive assortative
mating, the frequency of heterozygotes will decline and the frequency of homozygotes
will increase. Curiously, the gene frequencies won't change, so in the F1, f(A)
= .35 + 0.30/2 = 0.5... just as it was in the orginal population (f(A) = 0.2
+ 0.6/2 = 0.5). The genes are just being 'dealt' to offspring in a non-random
manner, affecting the genotypic frequencies at this locus.
So, suppose we observed the F1 population
in nature, and wanted to know if it was in HWE. We would calculate the gene
frequencies (A = 0.5, a = 0.5), and then estimate what the frequencies of the
genotypes would be IF the population was in HWE: p2 = 0.25, 2pq =
0.5, q2 = 0.25. We would compare our real population's genotypic
array with this HWE expectation, and see that they are not the same. And we
could see one thing more... we would see that the ACTUAL OBSERVED frequency
of heterozygotes (0.30) is LESS THAN the expected frequency of heterozygotes
under the HWE hypothesis (0.5). And we would see that the observed frequency
of both homozygotes is greater than expected. Knowing that positive assortative
mating can cause this pattern, we would have a working hypothesis regarding
the agent of evolution at work in this population.
b. Inbreeding: Mating with
a Relative
Inbreeding is mating with a relative.
It is similar to positive assortative mating, except that the two mates are
not just similar at one locus, but they are probably similar at MANY loci because
they are related and got their genes from the same ancestors. Siblings share,
on average, half their genes. Matings between siblings, then, will tend to reduce
heterozygosity at MANY loci, not just one.
The most extreme example is "obligate
self-fertilization". This is where a hermaphrodite ONLY mates with themselves.
This is not asexual reproduction - they produce gametes by meiosis and get all
the benefits of producing variable gametes that occurs in sexual reproduction;
but they only fertilize their own gametes. This has a profound effect on the
genetic structure of the population. Think about it: when an organism mates
with themselves, they are mating with an organism that has the SAME genotype
at EVERY locus. So, there will be a decrease in heterozygosity across the entire
genome, with a 50% reduction in heterozygosity each generation. This is the
most rapid loss possible. Siblings are only related, on average, by 50%, so
the loss of heterozygosity will only occur 1/2 as fast.... but it will still
occur at all loci across the genome.
Inbreeding often reduces reproductive
success, because there is an increase in homozygosity - and this means that
deleterious recessives are going to be expressed more frequently and exert their
negative effects on the offspring. A deleterious allele may be rare in a population,
but inbreeding will increase the probability that it occurs in the homozygous
condition and is expressed. Because inbreeding can reduce the survivorship of
offspring and thus reduce reproductive success of the parent, it is often selected
against. Selection favors different strategies that reduce the likelihood of
inbreeding, like "self-incompatibility" in some plants, like lions who push
male cubs out of a pride when they mature (thus they don't breed with their
sisters), or like humans who have a variety of cultural taboos against breeding
with relatives. However, inbreeding is also a mechanism for purging deleterious
alleles from the population. If a population can get through the first
few generations in which homozygote recessive are produced and selected against,
the net effect will be to eliminate these deleterious alleles from the population.
That can be a good thing in the long run.
4. Populations of Finite Size and Sampling Error/"Genetic
Drift"
The genetic structure of a population can change from generation to generation
just by chance - through a process statisticians call "sampling error"
and geneticists call "genetic drift". Think about it this way: suppose
we have a very large population in which p = 0.3 and q = 0.7, and the population
is in HWE to start; so the f(AA) = 0.09, f(Aa) = 0.21, and f(aa) = 0.49. Now,
suppose only 4 individuals mate, and suppose those four are just 'lucky' - they
don't mate because they are better adapted in any way, they just got lucky.
It is very unlikely that these four individuals will have the same genetic structure
as the whole population; small samples are notoriously unrepresentative and
variable. Indeed, all four may be 'aa', and the population will have changed
dramatically, losing the 'A' allele. This is why, to have confidence in any
observed pattern, you want a large sample. So, a population's genetic structure
may change just due to chance.
There are two important patterns
that result from this effect:
First, small samples will tend to
differ more from the original population than large samples. Second, since the
direction of this change is random, multiple small samples will tend to vary
more from one another, on average, than multiple large samples. So, small populations
will diverge more rapidly from one another, due to drift, than large populations.
There are two biological situations
where these effects are particularly important. The first is called a "Founder
Effect", where a small number of colonists establish a new population that
is isolated from the original population. Because this population of "founders"
is small, it is likely to differ dramatically from the original population....
and because it is small, it will also chance quickly just due to drift, as the
simulations in lab showed.
The second important instance in
which drift is important is called the "bottleneck effect". This is
when a large population is reduced in size. Typically this reduction is caused
by predation, a pathogen, or an environmental change. The survivors usually
make it because of selection at certain loci. For instance, survivors of a pathogenic
infection may be resistant to the pathogen. However, at other loci, the reduction
in population size causes genetic change due to drift.
So, CHANCE can be an agent of evolutionary
change. CHANCE, especially in small populations, can cause changes in
the genetic structure of a population. This is NOT selection - selection is
"non-random reproductive success" - the organisms that breed are "better" than
the others. Here, in Genetic Drift, the breeders are just LUCKY, not better.
It is random change. And, as the computer modes demonstrate, populations will
tend to become different genetically 'simply by chance', because these chance
changes are unlikely to be the SAME changes, or in the same direction, from
population to population. So, we can't STOP populations from evolving, really
- they will all change over time, even just do to random sampling error, and
they will tend to become different from other populations of the same species.
Of course, if the populations are very large, these random patterns of divergence
will be very slow.... but since no population is infintie in size, these random
changes will necessarily occur over time.
5. Natural Selection
1. Fitness Components:
As you know, natural selection is
"differential reproductive success" in a genetically variable population.
We can measure lifetime reproductive success as the number of successfully reproductive
offspring that a genotype produces, on average. This measureable quantity is called
"fitness". There are three factors that can influence 'fitness', or
reproductive success:
- difference in the probability of survival to reproductive age
- difference in the number of offspring produced
- difference in the probability that the OFFSPRING survive to reproductive
age (this is component of parental fitness, because the amount of energy that
a parent invests in the offspring can affect this probability)
Now, it seems like natural selection
would favor organisms that maximized all three components; and it would if that
were possible. However, organisms can't maximize all three components because
there are energetic constraints - organisms only have so much energy in their
'energy budget'. So, maximizing one component results in less energy that can
be invested in the other two. In addition, there are contradictory selective
pressures in the environment, such that a characteristic may increase fitness
with respect to one variable but decrease it with respect to another variable.
These are called 'trade-offs', and we will now examine these in more detail.
2. Constraints:
 a.
finite energy budgets and necessary trade-offs:
a.
finite energy budgets and necessary trade-offs:
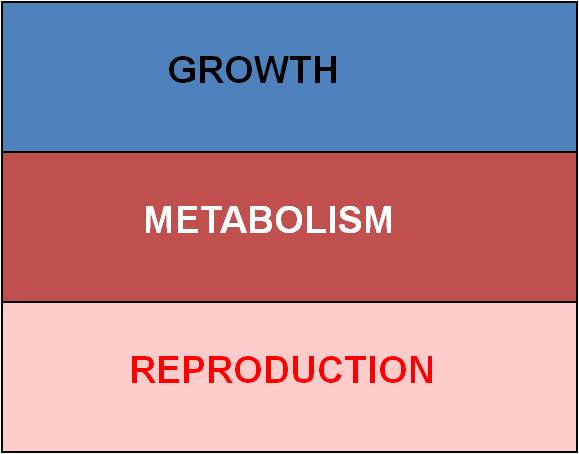
The most obvious constraint is ENERGY.
Every organism has a finite energy budget; it has harvested only so much energy
from the environment that it can allocate to all of its activities. There
are three major expenses:
- Basal metabolism - keeping the existing cells alive
- Growth - adding new cells, and keeping THEM alive (costs more energy above
basal metabolism)
- Reproduction - producing new cells and tissues, and raising offspring to
independence (costs even more energy).
So, with limited energy, increasing
one thing means that you must reduce costs in another area. These patterns
of energy allocation have direct effects on different fitness components.
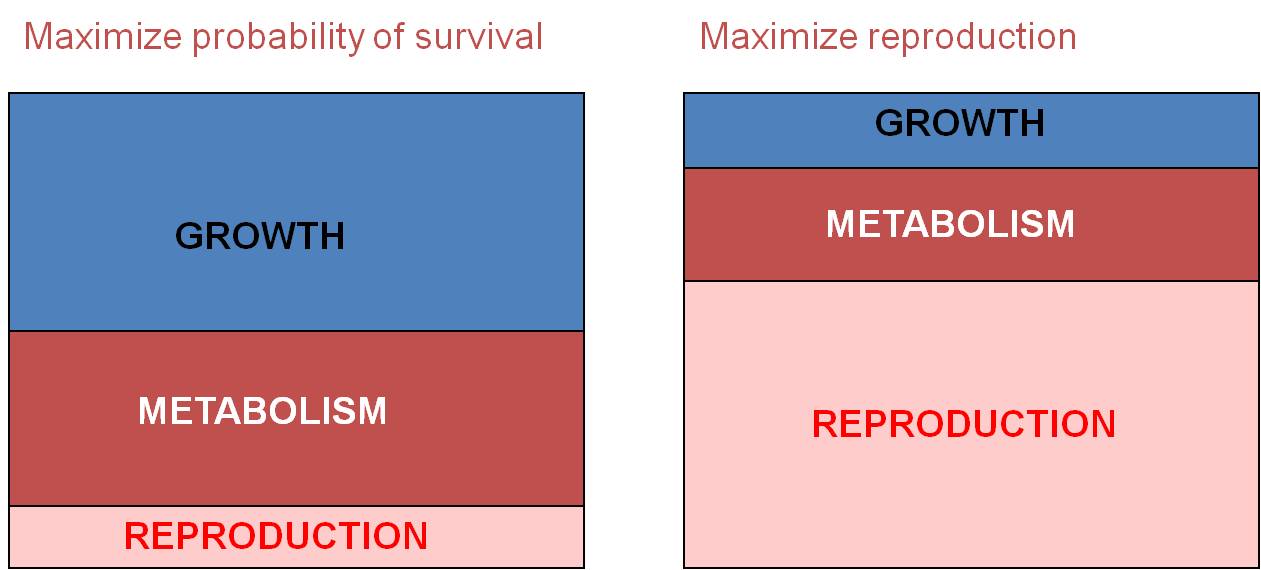 Trade-Off
#1: Survival vs. Immediate Reproduction: For example, ff an organism
maximizes energy investment in growth and metabolism, then they will increase
the probability that they survive. Why? Because being large tends to increase
the probability of survival, if only because there are fewer things that can
eat you. Likewise, large organisms are not as sensitive to changes in the environment.
Simply because of their larger size (and smaller surface area/volume ratio),
they don't lose heat, salt, water, or other materials to the environment as
rapidly as small organisms. However, by investing in growth, this means that
there is LESS energy to invest in immediate reproduction, meaning fewer offspring
can be produced. So, you CAN'T maximize all three components of selection
at the same time; selection seeks the best compromise in a given environment,
with particular biological potentials and constraints. Many organisms invest
in growth when young, to develop as rapidly as possible through these early,
vulnerable life-history stages. They delay reproduction completely when
they are young to maximize growth rate. Then, after they are older and
larger, they invest energy in reproduction (and growth rate SLOWS). These
organisms are "perennial" or "K" strategists - they are long-lived
organisms. Other organisms take a different strategy. They reproduce
early at the expense of growth. They don't survive long as a consequence
- they are "annual" or "r" strategists - species that live for less
than a year but invest almost all their energy in immediate reproduction. These
two examples are opposite sides of the same coin - they are both examples of
this same trade-off between survival and immediate reproduction; organism can't
maximize both at the same time, so they tend to maximize one or the other, or
change their pattern of allocation through their life.
Trade-Off
#1: Survival vs. Immediate Reproduction: For example, ff an organism
maximizes energy investment in growth and metabolism, then they will increase
the probability that they survive. Why? Because being large tends to increase
the probability of survival, if only because there are fewer things that can
eat you. Likewise, large organisms are not as sensitive to changes in the environment.
Simply because of their larger size (and smaller surface area/volume ratio),
they don't lose heat, salt, water, or other materials to the environment as
rapidly as small organisms. However, by investing in growth, this means that
there is LESS energy to invest in immediate reproduction, meaning fewer offspring
can be produced. So, you CAN'T maximize all three components of selection
at the same time; selection seeks the best compromise in a given environment,
with particular biological potentials and constraints. Many organisms invest
in growth when young, to develop as rapidly as possible through these early,
vulnerable life-history stages. They delay reproduction completely when
they are young to maximize growth rate. Then, after they are older and
larger, they invest energy in reproduction (and growth rate SLOWS). These
organisms are "perennial" or "K" strategists - they are long-lived
organisms. Other organisms take a different strategy. They reproduce
early at the expense of growth. They don't survive long as a consequence
- they are "annual" or "r" strategists - species that live for less
than a year but invest almost all their energy in immediate reproduction. These
two examples are opposite sides of the same coin - they are both examples of
this same trade-off between survival and immediate reproduction; organism can't
maximize both at the same time, so they tend to maximize one or the other, or
change their pattern of allocation through their life.
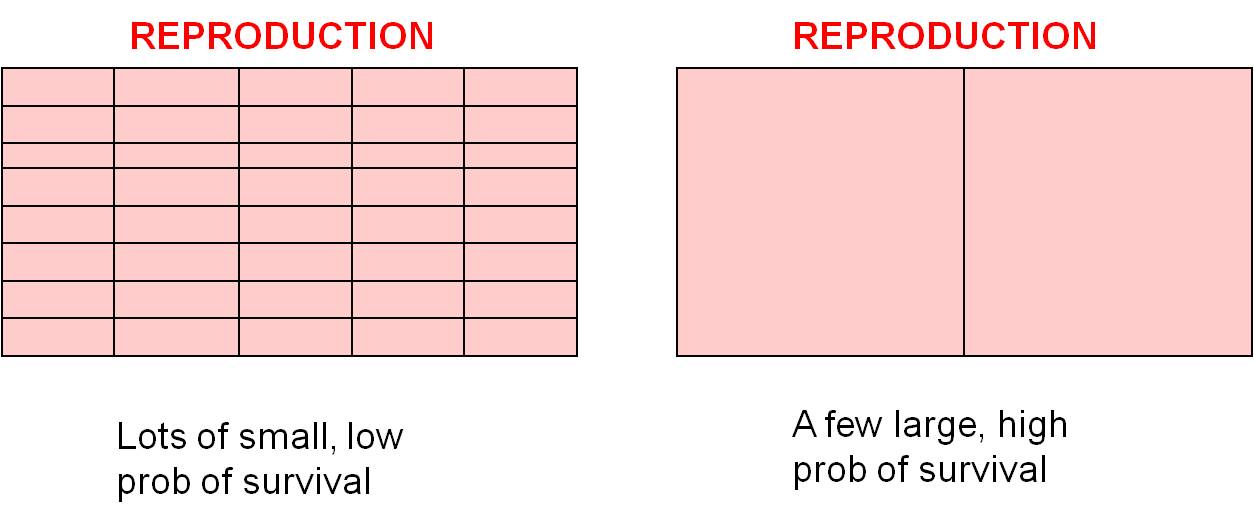 Trade
Off #2: Offspring Number vs. Offspring Size/Survivorship: There are
also trade-offs within the reproduction budget (the pink section in the budgets,
above). For the same "energetic cost", you can make lots of little offspring
or a few big offspring. As we discussed above, small organisms have a lower
probability of survival than large organisms, so you can make lots of small
offspring that each have a small chance of surviving, or you can invest in fewer
offspring and increase their chance of survival (by making them larger or by
investments in parent care). Small organisms, like insects, can't really make
a big offspring, so they exploit the other strategy and make lots of small offspring.
Large organisms can really take advantage of this 'choice', and selection will
favor different strategies in different large organisms. Some organisms
like mammals and birds tend to produce a few large offspring and invest significant
amounts of energy in parental care, further increasing the probability that
the offspring survive. Large vertebrates without parental care, like many
reptiles, produce more numerous smaller offspring and 'play the lottery' - increasing
the chance that one offspring survives by producing more offspring.
Trade
Off #2: Offspring Number vs. Offspring Size/Survivorship: There are
also trade-offs within the reproduction budget (the pink section in the budgets,
above). For the same "energetic cost", you can make lots of little offspring
or a few big offspring. As we discussed above, small organisms have a lower
probability of survival than large organisms, so you can make lots of small
offspring that each have a small chance of surviving, or you can invest in fewer
offspring and increase their chance of survival (by making them larger or by
investments in parent care). Small organisms, like insects, can't really make
a big offspring, so they exploit the other strategy and make lots of small offspring.
Large organisms can really take advantage of this 'choice', and selection will
favor different strategies in different large organisms. Some organisms
like mammals and birds tend to produce a few large offspring and invest significant
amounts of energy in parental care, further increasing the probability that
the offspring survive. Large vertebrates without parental care, like many
reptiles, produce more numerous smaller offspring and 'play the lottery' - increasing
the chance that one offspring survives by producing more offspring.
b.
Contradictory environmental pressures:
The environment is a complex place
- there are pressures that might favor some structures, and other pressures
acting AT THE SAME TIME that might select against that trait. Selection
can not maximize BOTH responses at the same time - so the solution will be a
suboptimal compromise. Consider leaf size. Big leaves are GOOD for
light absorption - they represent a larger solar panel that intercepts more
light. But big leaves are BAD for water loss - with a large SA/V ratio,
water is lost rapidly from a large thin leaf. So, the size of a leaf will
be a compromise solution to these two pressures... a solution that maximizes
neither function. The 'adaptive compromise' size depends on the relative
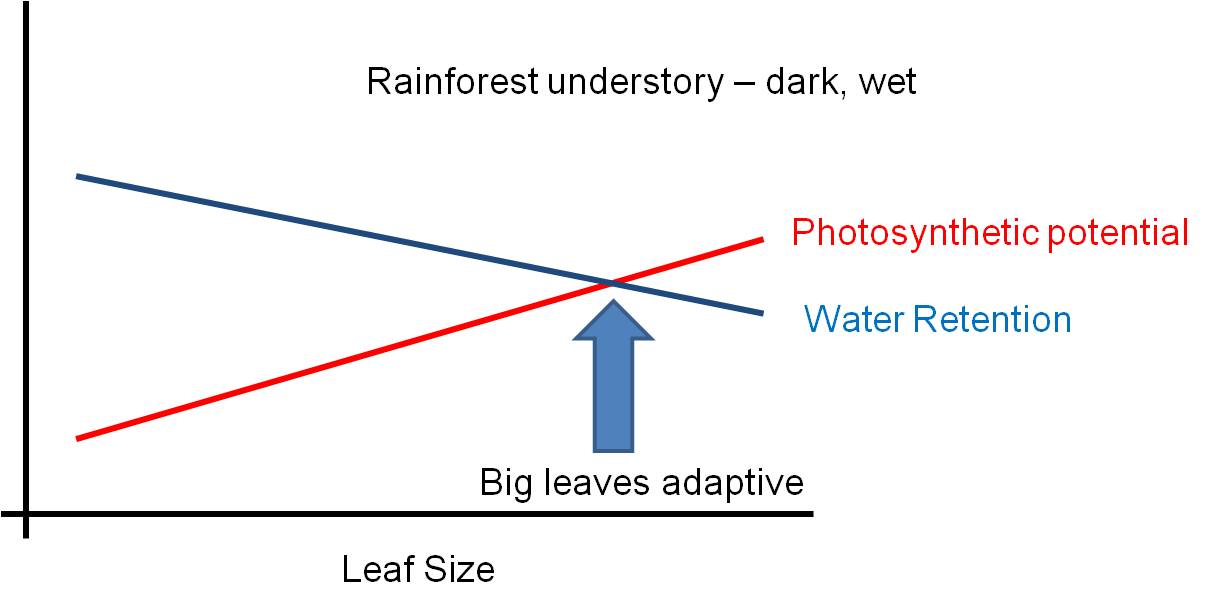
strengths of the contradictory pressures. If the risk of water loss is
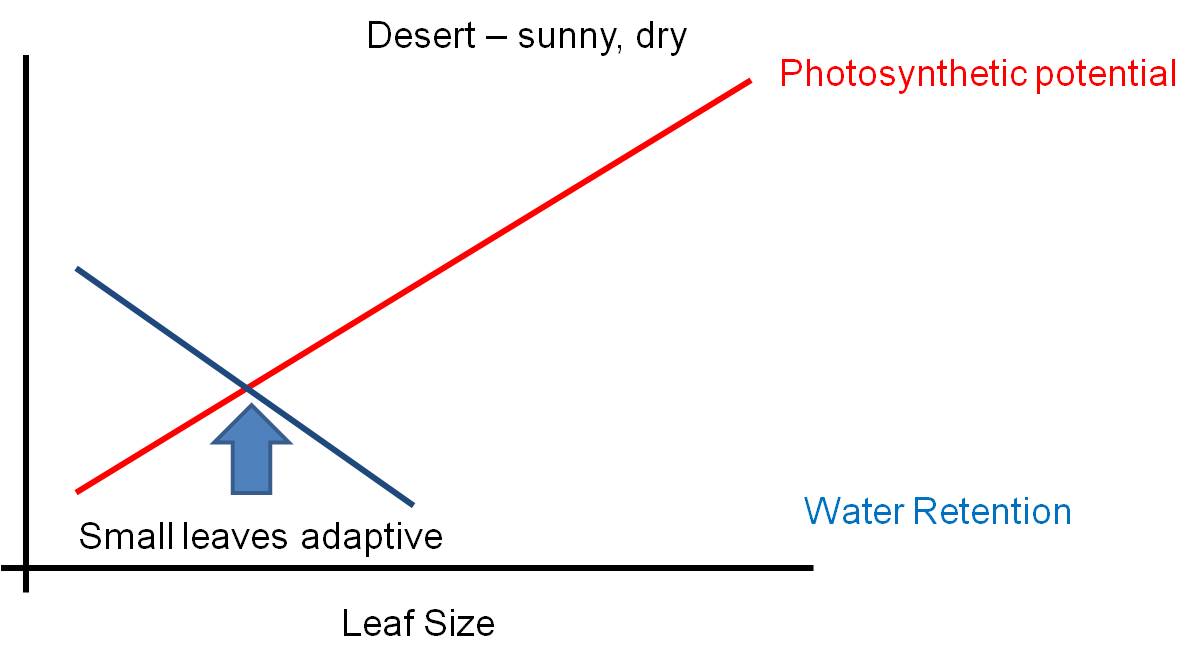
low (like in a rainforest), then the leaf can be large. If the risk of
water loss is high (like in the desert)and there is strong sun, then the selective
pressure to reduce leaf size is strong and leaves will be small or non-existent
(cacti).
For these reasons, 'perfect' adaptations
are impossible; there are biological costs to any adaptation (in terms of 'opportunity
costs' - NOT being able to do something else as well),and in terms of the complex
nature of the environment where selective pressures can be contradictory.
3. Modeling
Selection:
 a.
Calculating relative fitness:
a.
Calculating relative fitness:
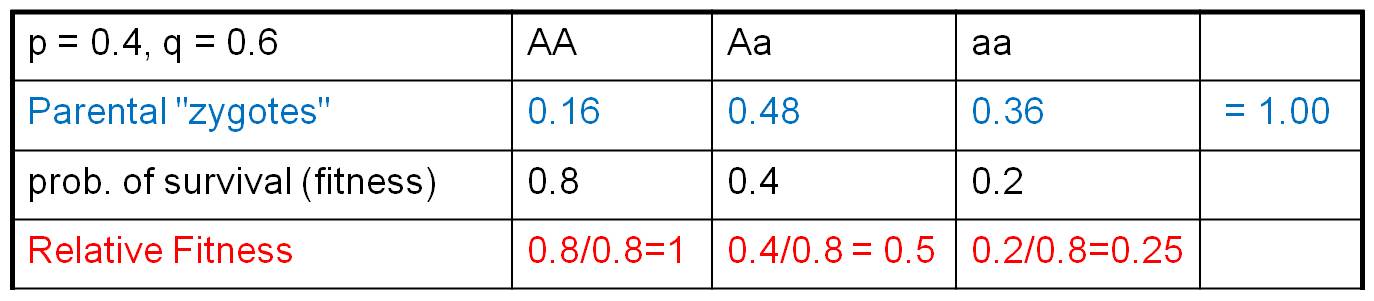
Consider the population to the right.
Suppose a population has JUST mated, creating parental zygotes in the genotypic
frequencies shown. Let's suppose that these zygotes, as a consequence of their
genotypes at this locus, have different probabilities of surviving to reproductive
age. But, to make things simple, lets assume that the other two components of
fitness are equal across the genotypes. So, differences in FITNESS are only
influenced by differences in the probability of survival to reproductive age.
Suppose those survival probabilities are 0.8, 0.4, and 0.2, respectively (as
shown).
These probabilities of survival are
'absolute' fitness values. Curiously, absolute fitness values are not very informative. What is important is relative reproductive success - reproductive success relative to the other genotypes
in the population. Think about it this way... suppose I tell you that "I
am thinking of an AA genotype that produces 1000 offspring a year. Do you think
the f(A) will increase or decrease in the population as a consequence of selection?"
Well, you might think, "wow, 1000 offspring is ALOT! Surely the f(A) will
increase!" But this would be an incorrect assumption. You need to know
what type of organism I am talking about, and how reproductively successful
the other genotypes are in the population. If this organism is a salmon, then
1000 offspring might be more than other genotypes in the population. But if
this organism is a clam, then the other genotypes might be producing 10's of
thousands of offspring - and the F(A) will decline. So, the key to selection
is differential reproductive success, which means
reproductive success relative to other organisms in the population. We represent
this as RELATIVE FITNESS, and calculate it by dividing all fitness values by
the LARGEST (ie., most FIT) value. This means that one genotype will necessarily
have a RELATIVE FITNESS = 1. In our case, above, we divide each fitness value
by the greatest value (0.8), so the relative fitness values are "1, 0.5,
and 0.25" for AA, Aa, and aa genotypes, respectively.
 b.
Modeling Selection:
b.
Modeling Selection:
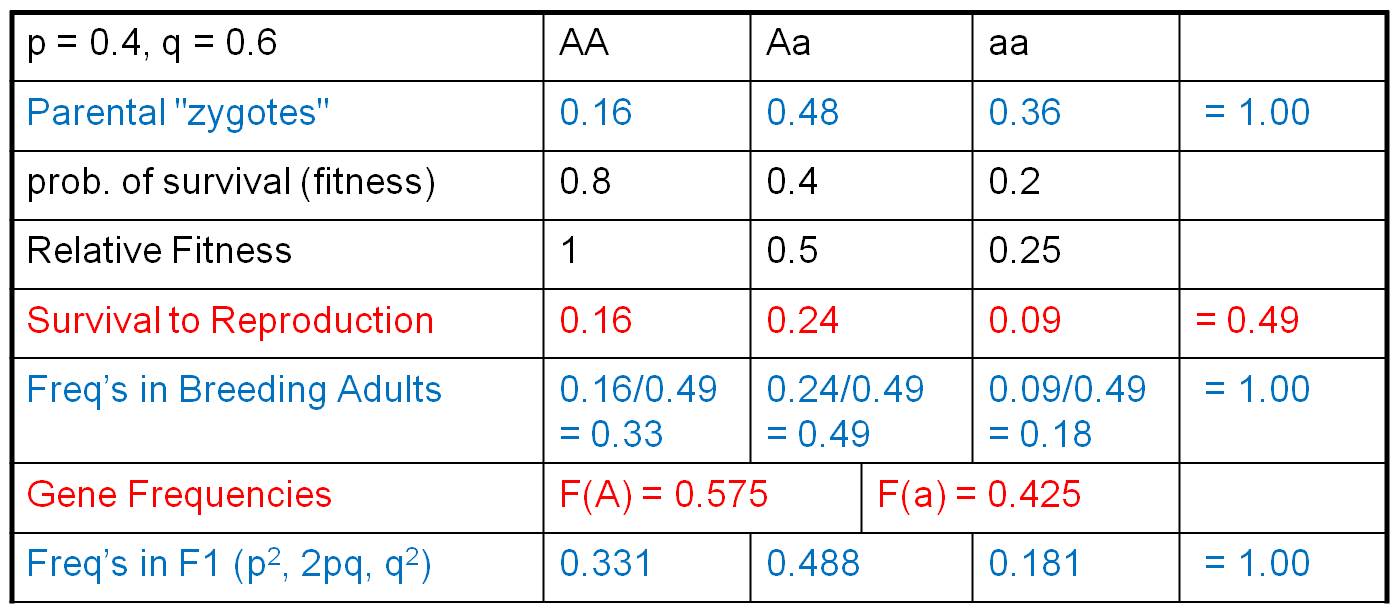
Now let's see what effect this selection
(in terms of differential survival) has on the genetic structure of the population.
First, we multiply the initial genotypic array by the relative fitness values.
For us, this describes the relative proportion of zygotes surviving to reproductive
age. So, these are the same organisms - we don't have a new generation yet -
it is just that the original zygotes have grown up and survived to adulthood
at different rates. Since many organisms have died, these genotypic values no
longer sum to 1. They sum to 0.49. If we want to know what the genotypic frequencies
are in the population of surviving reproductive adults (and WE DO), then we
must know what part of this new total is represented by each genotype. We calculate
that by dividing each genotypic value by the total (0.49), producing the genotypic
frequencies in this breeding population. These, of course, now sum to 1. From
here, we are home free. we calculate the gene frequencies in this reproductive
gene pool as:
F(A) = f(AA) = f(Aa)/2 = 0.575, and
f(a) = f(aa) + f(Aa)/2 = 0.425.
These sum to 1 so we've done the math correctly. Now, to produce the F1 zygotes,
assume that all other conditions of HWE are met (we are only modelling the direct
effect of selection, alone), so we assume that the organisms mate randomly and
calculate the genotypic frequencies of zygotes in the F1 using the terms: p2,
2pq, and q2.
So, as the result of differential
survival to reproductive age, A's and a's have not been transferred at the same
rate. The intial frequency of these genes was f(A) = 0.4 and f(a) = 0.6. After
one generation of differential reproductive success, the gene frequencies have
changed dramatically; now the 'A' gene is more abundant.
 4.
Types of Selection
4.
Types of Selection
Selection can act in a number of
ways in a population. Selection can favor one extreme phenotype over other phenotypes.
This is called "directional selection", and the mean phenotype in
the population (and the frequency of genes that influence the phenotype) should
move in one direction. There is also "stabilizing" selection, in which
an intermediate phenotype is most adaptive (think of sickle cell anemia in the
tropics, where the heterozygote had the greatest probability of survival and
reproduction). There is also "disruptive selection", in which both
extremes are more adaptive than the intermediate type. Although this may seem
unusual, it can be common for populations that exploit a complex environment.
One phenotype might work well in one microenvironment within that habitat, the
other extreme phenotype might work well in another specific microenvironment,
but the intermediate might not do well in either. Consider the pollination example
we used earlier for positive assortative mating. If there is a population of
four o'clocks living in a habitat with no diurnal pollinating insects, then
the pink flowers are selected against (don't reproduce) while the red and white
flowers are selected for.
Darwin recognized another very important
type of selection that he called "sexual selection". He realized that
some traits in certain species might DECREASE the probability of survival, and
yet might be adaptive. So, the long tail feathers in a peacock reduce survival:
they hinder the bird from flying, and they also make it really easy for predators
to see it. However, Darwin realized that these traits could still be adaptive
if the cost of decreased survival was outweighed by the gain in reproduction
while alive. So, Darwin appreciated the trade-offs that were possible in fitness
components. Competition among members of one sex for access to the other is
a type of sexual selection, too. When male bighorned sheep bash their heads
and fight, they reduce their probability of survival because they can get hurt.
However, winners increase their reproductive success so much that it compensates
for the cost of battle in terms of relative fitness.
E. Summary:
The period from 1900-1940 was a very
exciting and dynamic period in biology. With the rediscovery of Mendel's principles,
it seemed that biology had become a truly mathematical, predictive science.
The ability to predict patterns of heredity, and the transmission of 'mutant'
genes through generations, had a profound impact on evolutionary biology, too.
Many geneticists came to view mutation as the primary agent of evolutionary
change - not just as a primary source of variation. They viewed Darwin's ideas
of probabilistic Natural Selection as too weak to be responsible for the changes
seen in the fossil record. However, supporters of Darwin like Ernst
Mayr argued that random mutation could not explain the non-random adaptations
that were so obvious and pervasive in the natural world. The models of Hardy
and Weinberg, in the hands of new population geneticists like Theodosius
Dobzhansky and Sewall
Wright, were pivotal in resolving this dilemma. The resolution came in the
Modern Synthetic Theory of Evolution, developed by these scientists and others
in the 1930's and 1940's. As a consequence of the models you have just seen,
biologists realized that mutations were too rare to explain the changes seen
in natural populations over time. Rather, although mutation and recombination
were important as a source of new genetic variation, natural selection and genetic
drift were the primary agents that caused the genetic structure of population
to change over time. And so, as of 1940, our model of evolution looked like
this:
Things to Know:
1.
What are the five assumptions of the Hardy-Weinberg Equilibrium Model?
2.
Consider the following population:
| |
AA |
Aa |
aa |
| Number
of Individuals |
60 |
20 |
20 |
- -
calculate the genotypic frequencies.
- -
calculate the gene frequencies
- -
calculate the HARDY WEINBERG EQUILIBRIUM frequencies.
3.
If the HWE model does not describe any real population, how can it be useful?
1.
What are the five assumptions of the Hardy-Weinberg Equilibrium Model?
2.
Consider the following population:
| |
AA |
Aa |
aa |
| Number
of Individuals |
60 |
20 |
20 |
- -
calculate the genotypic frequencies.
- -
calculate the gene frequencies
- -
calculate the HARDY WEINBERG EQUILIBRIUM frequencies.
3.
If the HWE model does not describe any real population, how can it be useful?
Study Questions:
4.
Consider a population with p = 0.8 and q = 0.2. If the mutation rate of A-->
a = 4.0 x 10-6, what will the new gene frequencies be in the next
generation?
5.
Consider a population, p = 0.8 and q = 0.2. If migrants enter this population
with p = 0.1 and q = 0.9, such that immigrants comprise 15% of the total population,
what will the new gene frequencies be?
6. If the
population below undergoes positive assortative mating, what will the genotype
frequencies be in the next generation?
7. Why is
relative fitness more important than fitness?
8. Consider
the following population of zygotes:
AA
Aa
aa
Genotypic Frequency
0.3
0.3
0.4
Prob. of survival
0.4
0.2
0.1
a. What are
the initial gene frequencies?
b. Is the population
in HWE?
c. What are
the relative fitness values?
d. What are
the genotypic frequencies in the population of reproductive adults?
e. What are
the gene frequencies in the population of reproductive adults?
f. If there
is random mating, what will be the genotypic frequencies in the next generation?
g. What agent
of evolutionary change is at work?
9. Outline
the modern synthetic theory of evolution.
10. List the three
components of fitness, and explain two trade-offs that necessarily occur because
of limited energy budgets.
11. Why can selection
perfect an organism? Describe in terms of contradictory selective pressures,
and provide an example.
12.
How can the decimation of a population by overhunting cause changes in a population
due to both selection and drift?
 In
the early 20th century, at the same time that T. H. Morgan was studying mutations
and creating linkage maps, other biologists were considering the evolutionary
implications of this new knowledge regarding genetic variation. They appreciated
that individuals do not evolve - evolution is a process that occurs at the population
level. For example, as a consequence of differential reproductive success among
individuals in a population, the range of phenotypes and their relative frequencies
in the population will change over time. Individuals are born, life, reproduce
(maybe) and die. As a result of passing on their genes at different frequencies,
the genetic structure of the population changes over time (evolution). Two biologists,
G. Hardy and W. Weinberg, constructed a model to explain how the genetic structure
of a population might change over time.
In
the early 20th century, at the same time that T. H. Morgan was studying mutations
and creating linkage maps, other biologists were considering the evolutionary
implications of this new knowledge regarding genetic variation. They appreciated
that individuals do not evolve - evolution is a process that occurs at the population
level. For example, as a consequence of differential reproductive success among
individuals in a population, the range of phenotypes and their relative frequencies
in the population will change over time. Individuals are born, life, reproduce
(maybe) and die. As a result of passing on their genes at different frequencies,
the genetic structure of the population changes over time (evolution). Two biologists,
G. Hardy and W. Weinberg, constructed a model to explain how the genetic structure
of a population might change over time.
 The
easiest way to understand what these definitions represent is to work a problem
showing how they are computed.
The
easiest way to understand what these definitions represent is to work a problem
showing how they are computed.
 2.
Example:
2.
Example:
 -
Again, the coin analogy applies. No REAL coin is probably exactly perfectly
balanced. But, if I give you a coin and ask you how balanced it is, you flip
it a few times and compare its behavior to WHAT YOU WOULD EXPECT FROM A PERFECT
COIN (50:50 RATIO). Even though a perfectly balanced coin does not exist, we
can use this theoretical model as a benchmark, to compare the behavior of real
coins. Many real coins act in a manner that is consistent enough with
the expectations from a perfectly balanced coin that we are willing to use them
AS IF they were perfectly balanced. The Hardy Weinberg Equilibrium
Model is the same... it is a model of no change against which we can measure
real populations.
-
Again, the coin analogy applies. No REAL coin is probably exactly perfectly
balanced. But, if I give you a coin and ask you how balanced it is, you flip
it a few times and compare its behavior to WHAT YOU WOULD EXPECT FROM A PERFECT
COIN (50:50 RATIO). Even though a perfectly balanced coin does not exist, we
can use this theoretical model as a benchmark, to compare the behavior of real
coins. Many real coins act in a manner that is consistent enough with
the expectations from a perfectly balanced coin that we are willing to use them
AS IF they were perfectly balanced. The Hardy Weinberg Equilibrium
Model is the same... it is a model of no change against which we can measure
real populations.  3.
Non-Random Mating:
3.
Non-Random Mating:  a.
finite energy budgets and necessary trade-offs:
a.
finite energy budgets and necessary trade-offs: Trade-Off
#1: Survival vs. Immediate Reproduction: For example, ff an organism
maximizes energy investment in growth and metabolism, then they will increase
the probability that they survive. Why? Because being large tends to increase
the probability of survival, if only because there are fewer things that can
eat you. Likewise, large organisms are not as sensitive to changes in the environment.
Simply because of their larger size (and smaller surface area/volume ratio),
they don't lose heat, salt, water, or other materials to the environment as
rapidly as small organisms. However, by investing in growth, this means that
there is LESS energy to invest in immediate reproduction, meaning fewer offspring
can be produced. So, you CAN'T maximize all three components of selection
at the same time; selection seeks the best compromise in a given environment,
with particular biological potentials and constraints. Many organisms invest
in growth when young, to develop as rapidly as possible through these early,
vulnerable life-history stages. They delay reproduction completely when
they are young to maximize growth rate. Then, after they are older and
larger, they invest energy in reproduction (and growth rate SLOWS). These
organisms are "perennial" or "K" strategists - they are long-lived
organisms. Other organisms take a different strategy. They reproduce
early at the expense of growth. They don't survive long as a consequence
- they are "annual" or "r" strategists - species that live for less
than a year but invest almost all their energy in immediate reproduction. These
two examples are opposite sides of the same coin - they are both examples of
this same trade-off between survival and immediate reproduction; organism can't
maximize both at the same time, so they tend to maximize one or the other, or
change their pattern of allocation through their life.
Trade-Off
#1: Survival vs. Immediate Reproduction: For example, ff an organism
maximizes energy investment in growth and metabolism, then they will increase
the probability that they survive. Why? Because being large tends to increase
the probability of survival, if only because there are fewer things that can
eat you. Likewise, large organisms are not as sensitive to changes in the environment.
Simply because of their larger size (and smaller surface area/volume ratio),
they don't lose heat, salt, water, or other materials to the environment as
rapidly as small organisms. However, by investing in growth, this means that
there is LESS energy to invest in immediate reproduction, meaning fewer offspring
can be produced. So, you CAN'T maximize all three components of selection
at the same time; selection seeks the best compromise in a given environment,
with particular biological potentials and constraints. Many organisms invest
in growth when young, to develop as rapidly as possible through these early,
vulnerable life-history stages. They delay reproduction completely when
they are young to maximize growth rate. Then, after they are older and
larger, they invest energy in reproduction (and growth rate SLOWS). These
organisms are "perennial" or "K" strategists - they are long-lived
organisms. Other organisms take a different strategy. They reproduce
early at the expense of growth. They don't survive long as a consequence
- they are "annual" or "r" strategists - species that live for less
than a year but invest almost all their energy in immediate reproduction. These
two examples are opposite sides of the same coin - they are both examples of
this same trade-off between survival and immediate reproduction; organism can't
maximize both at the same time, so they tend to maximize one or the other, or
change their pattern of allocation through their life. Trade
Off #2: Offspring Number vs. Offspring Size/Survivorship: There are
also trade-offs within the reproduction budget (the pink section in the budgets,
above). For the same "energetic cost", you can make lots of little offspring
or a few big offspring. As we discussed above, small organisms have a lower
probability of survival than large organisms, so you can make lots of small
offspring that each have a small chance of surviving, or you can invest in fewer
offspring and increase their chance of survival (by making them larger or by
investments in parent care). Small organisms, like insects, can't really make
a big offspring, so they exploit the other strategy and make lots of small offspring.
Large organisms can really take advantage of this 'choice', and selection will
favor different strategies in different large organisms. Some organisms
like mammals and birds tend to produce a few large offspring and invest significant
amounts of energy in parental care, further increasing the probability that
the offspring survive. Large vertebrates without parental care, like many
reptiles, produce more numerous smaller offspring and 'play the lottery' - increasing
the chance that one offspring survives by producing more offspring.
Trade
Off #2: Offspring Number vs. Offspring Size/Survivorship: There are
also trade-offs within the reproduction budget (the pink section in the budgets,
above). For the same "energetic cost", you can make lots of little offspring
or a few big offspring. As we discussed above, small organisms have a lower
probability of survival than large organisms, so you can make lots of small
offspring that each have a small chance of surviving, or you can invest in fewer
offspring and increase their chance of survival (by making them larger or by
investments in parent care). Small organisms, like insects, can't really make
a big offspring, so they exploit the other strategy and make lots of small offspring.
Large organisms can really take advantage of this 'choice', and selection will
favor different strategies in different large organisms. Some organisms
like mammals and birds tend to produce a few large offspring and invest significant
amounts of energy in parental care, further increasing the probability that
the offspring survive. Large vertebrates without parental care, like many
reptiles, produce more numerous smaller offspring and 'play the lottery' - increasing
the chance that one offspring survives by producing more offspring. 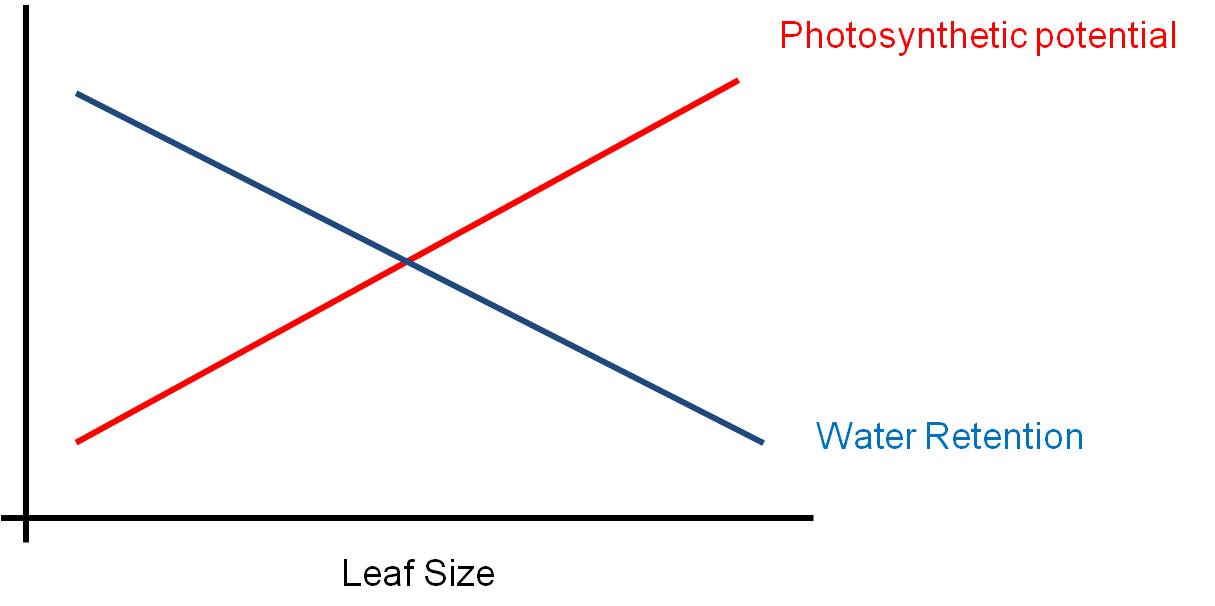
 a.
Calculating relative fitness:
a.
Calculating relative fitness: b.
Modeling Selection:
b.
Modeling Selection: 4.
Types of Selection
4.
Types of Selection